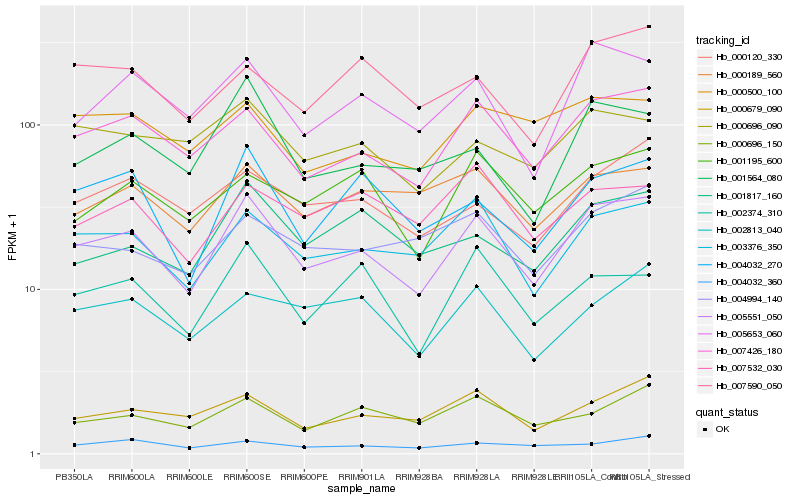
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005551_050 |
0.0 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 2 |
Hb_007426_180 |
0.076798747 |
- |
- |
PREDICTED: probable inositol transporter 2 [Jatropha curcas] |
| 3 |
Hb_003376_350 |
0.0898099677 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005653_060 |
0.1011994325 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 5 |
Hb_000679_090 |
0.1092388001 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |
| 6 |
Hb_001564_080 |
0.1119149115 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 7 |
Hb_004032_270 |
0.1147235996 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000120_330 |
0.1158895302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004994_140 |
0.116429629 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002374_310 |
0.1167808202 |
- |
- |
PREDICTED: uncharacterized protein LOC105638955 [Jatropha curcas] |
| 11 |
Hb_001817_160 |
0.1175017896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000189_560 |
0.1179731032 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1 [Jatropha curcas] |
| 13 |
Hb_001195_600 |
0.1179992111 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 14 |
Hb_002813_040 |
0.1186826796 |
- |
- |
PREDICTED: PRA1 family protein G2 [Jatropha curcas] |
| 15 |
Hb_000696_150 |
0.119455495 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g51320 [Jatropha curcas] |
| 16 |
Hb_000696_090 |
0.1214696541 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |
| 17 |
Hb_007590_050 |
0.1218856587 |
- |
- |
NudC domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000500_100 |
0.1224056646 |
- |
- |
peptide methionine sulfoxide reductase A1-like [Jatropha curcas] |
| 19 |
Hb_004032_360 |
0.1229424226 |
- |
- |
PREDICTED: uncharacterized protein LOC103403885 [Malus domestica] |
| 20 |
Hb_007532_030 |
0.12512458 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |