| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
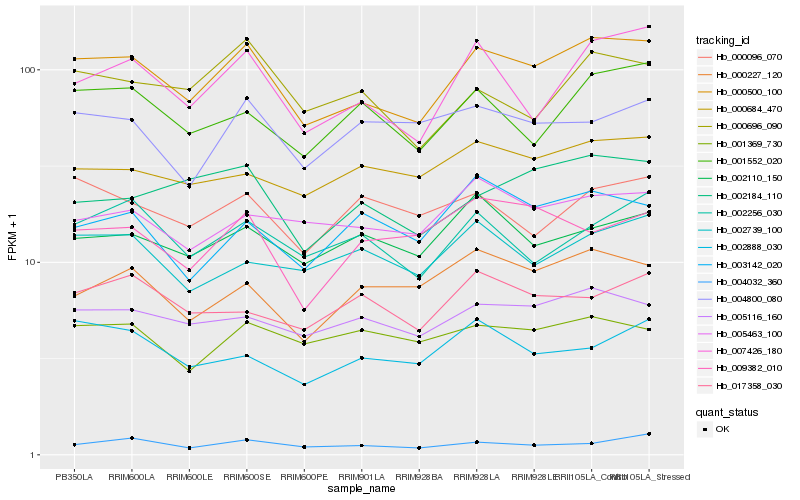
Hb_000500_100 |
0.0 |
- |
- |
peptide methionine sulfoxide reductase A1-like [Jatropha curcas] |
| 2 |
Hb_007426_180 |
0.0872826723 |
- |
- |
PREDICTED: probable inositol transporter 2 [Jatropha curcas] |
| 3 |
Hb_005116_160 |
0.0903380576 |
transcription factor |
TF Family: TRAF |
- |
| 4 |
Hb_001552_020 |
0.0994625052 |
- |
- |
PREDICTED: uncharacterized protein LOC105639653 [Jatropha curcas] |
| 5 |
Hb_002184_110 |
0.1000355609 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 [Jatropha curcas] |
| 6 |
Hb_005463_100 |
0.1007767914 |
- |
- |
hypothetical protein POPTR_0001s08400g [Populus trichocarpa] |
| 7 |
Hb_002256_030 |
0.1010324319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000684_470 |
0.1014449358 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 9 |
Hb_002110_150 |
0.1018311584 |
- |
- |
PREDICTED: serine/threonine-protein kinase PEPKR2 [Jatropha curcas] |
| 10 |
Hb_000696_090 |
0.1025865713 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |
| 11 |
Hb_000227_120 |
0.1030423761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004032_360 |
0.1045486556 |
- |
- |
PREDICTED: uncharacterized protein LOC103403885 [Malus domestica] |
| 13 |
Hb_002739_100 |
0.1049936946 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002888_030 |
0.1077614128 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 15 |
Hb_001369_730 |
0.1079777014 |
- |
- |
F5O11.10, putative isoform 1 [Theobroma cacao] |
| 16 |
Hb_017358_030 |
0.1090333792 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000096_070 |
0.1090708753 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 18 |
Hb_004800_080 |
0.1091463998 |
- |
- |
PREDICTED: acidic leucine-rich nuclear phosphoprotein 32-related protein [Jatropha curcas] |
| 19 |
Hb_003142_020 |
0.1093445744 |
- |
- |
PREDICTED: uncharacterized protein LOC105648334 [Jatropha curcas] |
| 20 |
Hb_009382_010 |
0.1098575816 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |