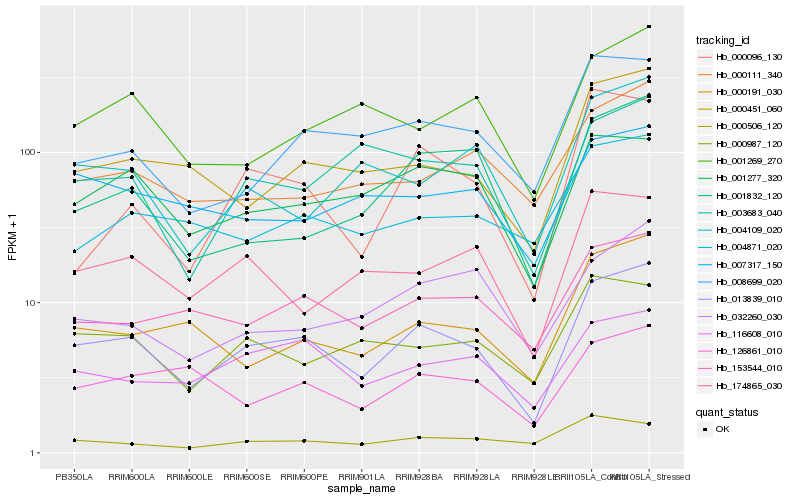
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013839_010 |
0.0 |
- |
- |
Angio-associated migratory cell protein, putative [Ricinus communis] |
| 2 |
Hb_000451_060 |
0.1271825667 |
- |
- |
hypothetical protein CICLE_v10010074mg [Citrus clementina] |
| 3 |
Hb_153544_010 |
0.1348667333 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Populus euphratica] |
| 4 |
Hb_000191_030 |
0.140835544 |
- |
- |
hypothetical protein POPTR_0012s07830g [Populus trichocarpa] |
| 5 |
Hb_008699_020 |
0.1432149491 |
- |
- |
eukaryotic translation initiation factor 5A isoform IV [Hevea brasiliensis] |
| 6 |
Hb_116608_010 |
0.1458968124 |
transcription factor |
TF Family: NF-YC |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_001277_320 |
0.1478115869 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 8 |
Hb_000987_120 |
0.1486729444 |
- |
- |
PREDICTED: protein SIEL [Jatropha curcas] |
| 9 |
Hb_174865_030 |
0.1489221547 |
- |
- |
PREDICTED: hemiasterlin resistant protein 1 [Eucalyptus grandis] |
| 10 |
Hb_004871_020 |
0.1505115143 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 11 |
Hb_000111_340 |
0.1514345568 |
- |
- |
peptidyl-prolyl cis-trans isomerase h, ppih, putative [Ricinus communis] |
| 12 |
Hb_001269_270 |
0.1521737784 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 13 |
Hb_001832_120 |
0.1545022877 |
- |
- |
hypothetical protein PHAVU_003G136800g [Phaseolus vulgaris] |
| 14 |
Hb_003683_040 |
0.1557438271 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_004109_020 |
0.1561801799 |
- |
- |
60S ribosomal protein L18aA [Hevea brasiliensis] |
| 16 |
Hb_000506_120 |
0.1568967747 |
- |
- |
hypothetical protein AALP_AA6G154200 [Arabis alpina] |
| 17 |
Hb_000096_130 |
0.1572504333 |
- |
- |
hypothetical protein 23 [Hevea brasiliensis] |
| 18 |
Hb_126861_010 |
0.1575565277 |
- |
- |
hypothetical protein M569_05914, partial [Genlisea aurea] |
| 19 |
Hb_007317_150 |
0.1575984052 |
- |
- |
PREDICTED: 40S ribosomal protein S25 [Jatropha curcas] |
| 20 |
Hb_032260_030 |
0.157882792 |
- |
- |
hypothetical protein POPTR_0001s10500g [Populus trichocarpa] |