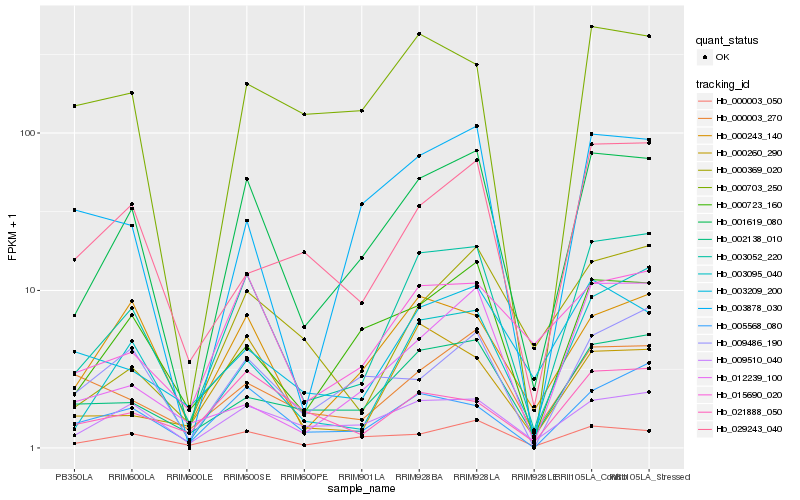
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003052_220 |
0.0 |
- |
- |
- |
| 2 |
Hb_001619_080 |
0.098363278 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003095_040 |
0.1365350092 |
- |
- |
hypothetical protein CICLE_v10030016mg [Citrus clementina] |
| 4 |
Hb_002138_010 |
0.1605555989 |
- |
- |
hypothetical protein POPTR_0001s30620g [Populus trichocarpa] |
| 5 |
Hb_005568_080 |
0.17516085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000260_290 |
0.1852427442 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_015690_020 |
0.1859315487 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 8 |
Hb_000243_140 |
0.1875849687 |
transcription factor |
TF Family: SRS |
Lateral root primordium protein-related, putative [Theobroma cacao] |
| 9 |
Hb_000723_160 |
0.1890794323 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP11 [Jatropha curcas] |
| 10 |
Hb_009510_040 |
0.1951727965 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen2-1-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_029243_040 |
0.202576358 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 2, mitochondrial-like [Populus euphratica] |
| 12 |
Hb_003878_030 |
0.2028735599 |
- |
- |
hypothetical protein POPTR_0011s11120g [Populus trichocarpa] |
| 13 |
Hb_000369_020 |
0.2072722849 |
- |
- |
PREDICTED: probable fructokinase-7 [Jatropha curcas] |
| 14 |
Hb_000703_250 |
0.2077319288 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 10-like [Jatropha curcas] |
| 15 |
Hb_000003_050 |
0.208686106 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 16 |
Hb_000003_270 |
0.2098899655 |
- |
- |
PREDICTED: cytokinin dehydrogenase 6 [Jatropha curcas] |
| 17 |
Hb_003209_200 |
0.2110894215 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1621230 [Ricinus communis] |
| 18 |
Hb_012239_100 |
0.2123080675 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 19 |
Hb_009486_190 |
0.2138207849 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_021888_050 |
0.2152681858 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |