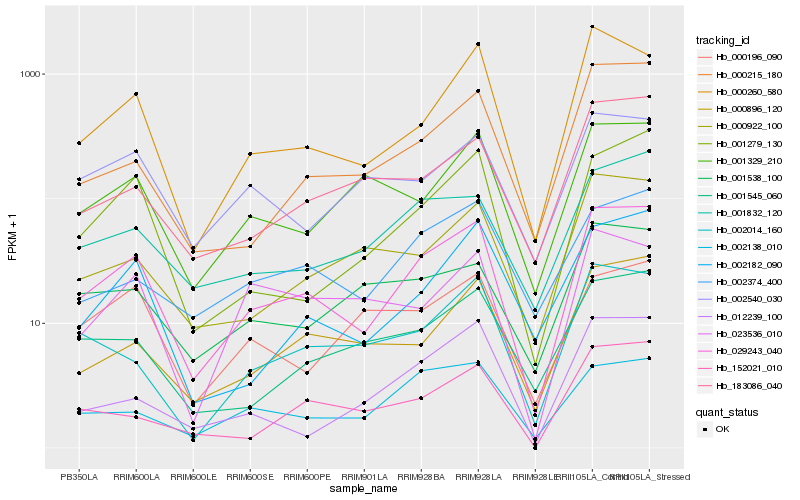
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029243_040 |
0.0 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 2, mitochondrial-like [Populus euphratica] |
| 2 |
Hb_000260_580 |
0.1214283868 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001279_130 |
0.1369191444 |
- |
- |
hypothetical protein L484_017676 [Morus notabilis] |
| 4 |
Hb_000896_120 |
0.1434614425 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 5 |
Hb_001832_120 |
0.1436735369 |
- |
- |
hypothetical protein PHAVU_003G136800g [Phaseolus vulgaris] |
| 6 |
Hb_000215_180 |
0.1438710992 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 7 |
Hb_002182_090 |
0.1449676171 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF4.6-like isoform X1 [Gossypium raimondii] |
| 8 |
Hb_002138_010 |
0.1487246168 |
- |
- |
hypothetical protein POPTR_0001s30620g [Populus trichocarpa] |
| 9 |
Hb_001545_060 |
0.1502936522 |
- |
- |
PREDICTED: uncharacterized protein LOC105643880 [Jatropha curcas] |
| 10 |
Hb_002014_160 |
0.150404201 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor C-1 [Jatropha curcas] |
| 11 |
Hb_002374_400 |
0.1508786596 |
transcription factor |
TF Family: TRAF |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_000922_100 |
0.1530863912 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 13 |
Hb_001329_210 |
0.1559352771 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 14 |
Hb_183086_040 |
0.157746221 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |
| 15 |
Hb_152021_010 |
0.1599258538 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002540_030 |
0.1636608611 |
- |
- |
hypothetical protein 32 [Hevea brasiliensis] |
| 17 |
Hb_001538_100 |
0.1654906935 |
- |
- |
PREDICTED: histone deacetylase 6 [Jatropha curcas] |
| 18 |
Hb_012239_100 |
0.1705304502 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 19 |
Hb_023536_010 |
0.1711169053 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 1 [Jatropha curcas] |
| 20 |
Hb_000196_090 |
0.1726406088 |
- |
- |
axonemal dynein light chain, putative [Ricinus communis] |