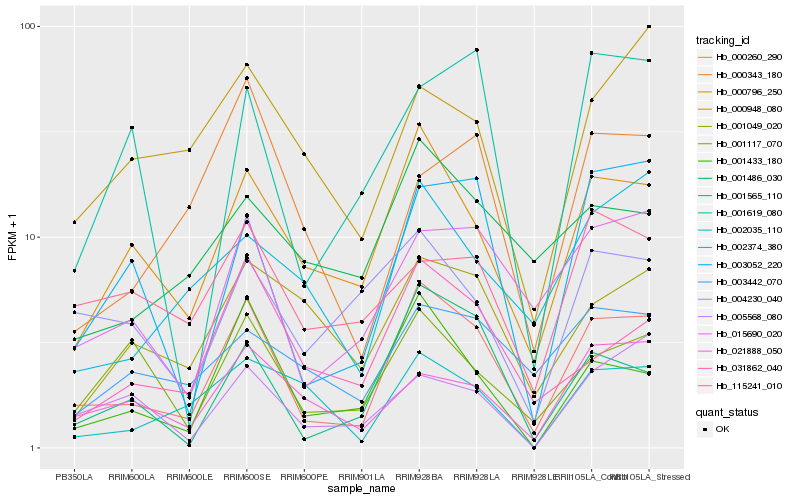
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_290 |
0.0 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000796_250 |
0.1582907011 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 3 |
Hb_015690_020 |
0.1746849347 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 4 |
Hb_001433_180 |
0.1802929662 |
- |
- |
hypothetical protein JCGZ_23261 [Jatropha curcas] |
| 5 |
Hb_001565_110 |
0.1837588335 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 6 |
Hb_003052_220 |
0.1852427442 |
- |
- |
- |
| 7 |
Hb_031862_040 |
0.1943393398 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 8 |
Hb_001117_070 |
0.2044732658 |
- |
- |
30S ribosomal protein S19, putative [Ricinus communis] |
| 9 |
Hb_021888_050 |
0.2049247006 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 10 |
Hb_002374_380 |
0.2150128752 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_001049_020 |
0.2172901505 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004230_040 |
0.2178213298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003442_070 |
0.2182907091 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 14 |
Hb_002035_110 |
0.2208501266 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 15 |
Hb_001486_030 |
0.2214634616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005568_080 |
0.2229837152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001619_080 |
0.2251319919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000343_180 |
0.227867779 |
- |
- |
NDWp3 [Podospora anserina] |
| 19 |
Hb_115241_010 |
0.2287514765 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Jatropha curcas] |
| 20 |
Hb_000948_080 |
0.2290620807 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |