| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
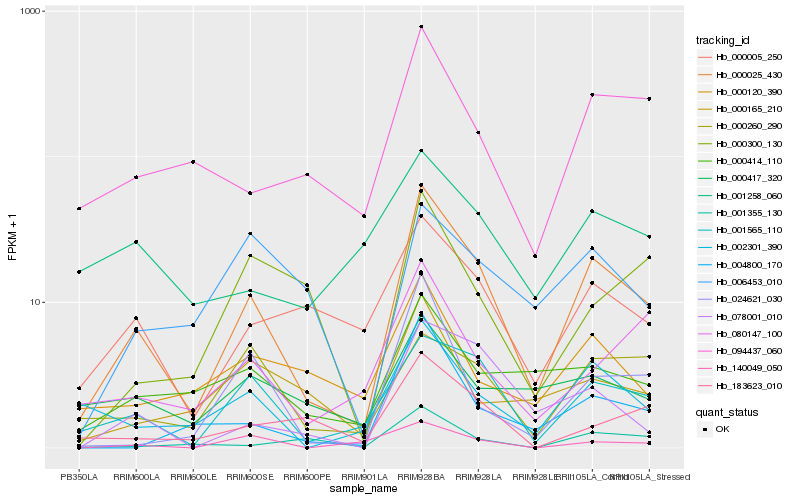
Hb_000025_430 |
0.0 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_001565_110 |
0.1975039309 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 3 |
Hb_002301_390 |
0.2239181619 |
- |
- |
- |
| 4 |
Hb_078001_010 |
0.2388747276 |
- |
- |
Uncharacterized protein TCM_000398 [Theobroma cacao] |
| 5 |
Hb_000005_250 |
0.2400098511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_094437_060 |
0.2406299744 |
- |
- |
PREDICTED: cytochrome B5 isoform D-like [Jatropha curcas] |
| 7 |
Hb_004800_170 |
0.2410308482 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 8 |
Hb_024621_030 |
0.2457767303 |
- |
- |
tetraketide alpha-pyrone reductase 2 [Jatropha curcas] |
| 9 |
Hb_000414_110 |
0.2540906654 |
- |
- |
PREDICTED: MATE efflux family protein 5 isoform X1 [Populus euphratica] |
| 10 |
Hb_080147_100 |
0.2562740902 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN [Jatropha curcas] |
| 11 |
Hb_183623_010 |
0.2597590172 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 12 |
Hb_001355_130 |
0.2613189858 |
- |
- |
PREDICTED: uncharacterized protein LOC105137896 [Populus euphratica] |
| 13 |
Hb_000165_210 |
0.2654521416 |
- |
- |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000120_390 |
0.2674387274 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 15 |
Hb_140049_050 |
0.2710221497 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 16 |
Hb_000260_290 |
0.2747160155 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000417_320 |
0.2784032971 |
- |
- |
hypothetical protein CICLE_v10000333mg [Citrus clementina] |
| 18 |
Hb_000300_130 |
0.2793656447 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 19 |
Hb_006453_010 |
0.2797546484 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 20 |
Hb_001258_060 |
0.2815599067 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |