| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
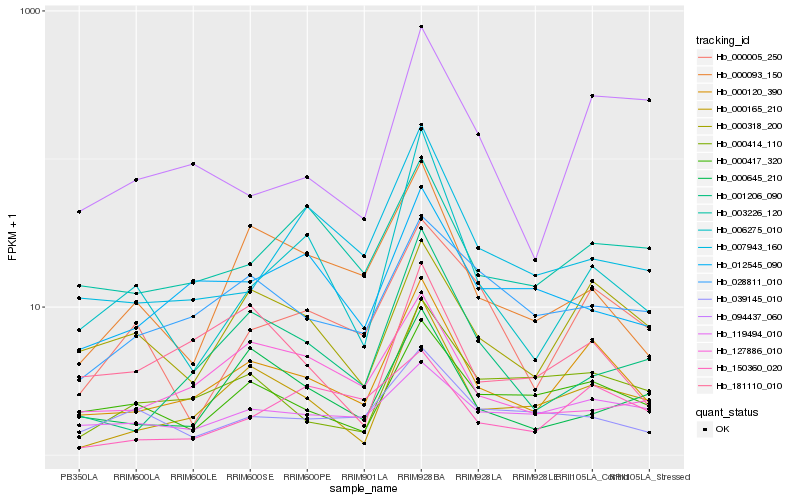
Hb_000120_390 |
0.0 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 2 |
Hb_000165_210 |
0.1570060247 |
- |
- |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000318_200 |
0.1607534507 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 4 |
Hb_000417_320 |
0.1673617268 |
- |
- |
hypothetical protein CICLE_v10000333mg [Citrus clementina] |
| 5 |
Hb_000414_110 |
0.1887531083 |
- |
- |
PREDICTED: MATE efflux family protein 5 isoform X1 [Populus euphratica] |
| 6 |
Hb_181110_010 |
0.1900194643 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 7 |
Hb_003226_120 |
0.1911629535 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000093_150 |
0.1938595728 |
- |
- |
6-phosphogluconolactonase, putative [Ricinus communis] |
| 9 |
Hb_028811_010 |
0.198948601 |
- |
- |
PREDICTED: uncharacterized protein LOC105650178 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001206_090 |
0.1998990334 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 11 |
Hb_000005_250 |
0.2014769132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007943_160 |
0.20152827 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 13 |
Hb_012545_090 |
0.2016758364 |
- |
- |
hypothetical protein JCGZ_04320 [Jatropha curcas] |
| 14 |
Hb_119494_010 |
0.2036560424 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 15 |
Hb_000645_210 |
0.208581762 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_150360_020 |
0.2114437371 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 17 |
Hb_127886_010 |
0.2115429874 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 18 |
Hb_094437_060 |
0.2116408633 |
- |
- |
PREDICTED: cytochrome B5 isoform D-like [Jatropha curcas] |
| 19 |
Hb_006275_010 |
0.211916611 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 20 |
Hb_039145_010 |
0.2121292337 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |