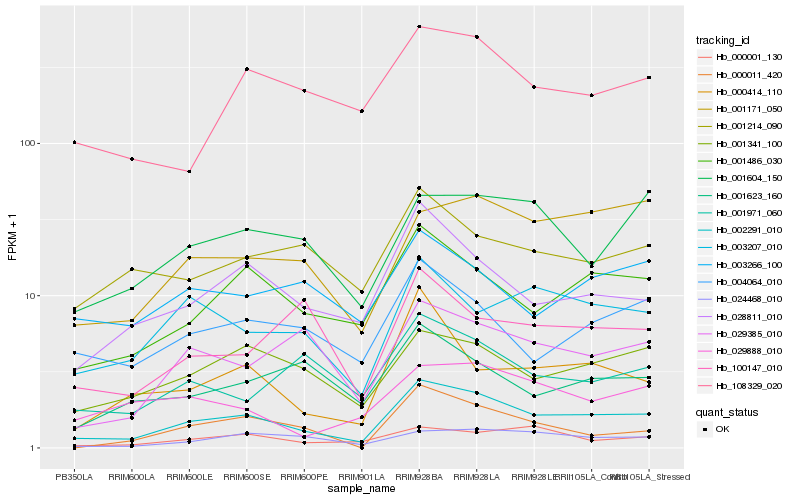
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002291_010 |
0.0 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 2 |
Hb_001486_030 |
0.1465896314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_028811_010 |
0.1532388072 |
- |
- |
PREDICTED: uncharacterized protein LOC105650178 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001341_100 |
0.1583262395 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |
| 5 |
Hb_003207_010 |
0.166483868 |
- |
- |
PREDICTED: phospholipase D beta 1-like [Jatropha curcas] |
| 6 |
Hb_001171_050 |
0.1687821745 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0284459 [Jatropha curcas] |
| 7 |
Hb_004064_010 |
0.1714345221 |
- |
- |
PREDICTED: uncharacterized protein LOC105133340 isoform X1 [Populus euphratica] |
| 8 |
Hb_029385_010 |
0.1723552721 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |
| 9 |
Hb_108329_020 |
0.1733033074 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 10 |
Hb_024468_010 |
0.1748611199 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 11 |
Hb_001604_150 |
0.1812566679 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Nelumbo nucifera] |
| 12 |
Hb_001971_060 |
0.1830094662 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Populus euphratica] |
| 13 |
Hb_000414_110 |
0.1845258968 |
- |
- |
PREDICTED: MATE efflux family protein 5 isoform X1 [Populus euphratica] |
| 14 |
Hb_000001_130 |
0.1851008772 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 15 |
Hb_001214_090 |
0.1862698023 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 16 |
Hb_000011_420 |
0.1884538026 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g29660-like [Citrus sinensis] |
| 17 |
Hb_029888_010 |
0.1904399484 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Nicotiana tomentosiformis] |
| 18 |
Hb_001623_160 |
0.1908616245 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 19 |
Hb_100147_010 |
0.1916712108 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 20 |
Hb_003266_100 |
0.1927633497 |
- |
- |
Isomerase protein [Gossypium arboreum] |