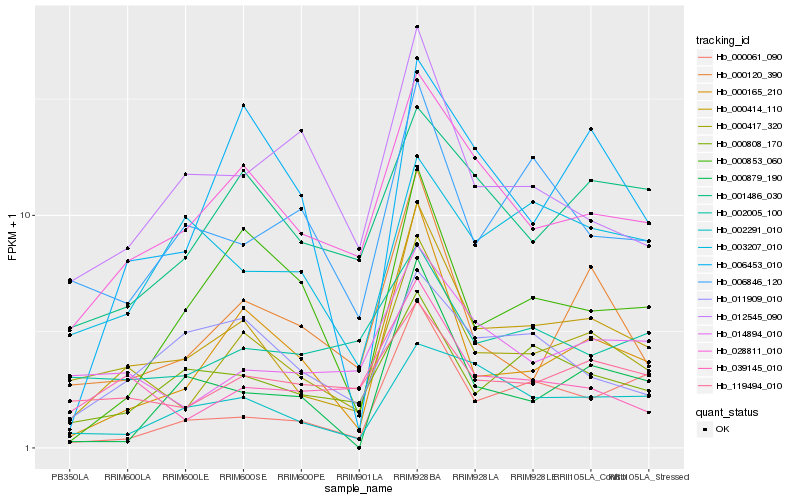
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000414_110 |
0.0 |
- |
- |
PREDICTED: MATE efflux family protein 5 isoform X1 [Populus euphratica] |
| 2 |
Hb_000417_320 |
0.1341289218 |
- |
- |
hypothetical protein CICLE_v10000333mg [Citrus clementina] |
| 3 |
Hb_028811_010 |
0.1453787892 |
- |
- |
PREDICTED: uncharacterized protein LOC105650178 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000165_210 |
0.1614764891 |
- |
- |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000808_170 |
0.1668640993 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 6 |
Hb_000061_090 |
0.1749405044 |
- |
- |
hypothetical protein B456_003G0981001, partial [Gossypium raimondii] |
| 7 |
Hb_002291_010 |
0.1845258968 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_000120_390 |
0.1887531083 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 9 |
Hb_001486_030 |
0.1903039853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006846_120 |
0.1933700722 |
- |
- |
Succinate/fumarate mitochondrial transporter, putative [Ricinus communis] |
| 11 |
Hb_011909_010 |
0.1958863819 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |
| 12 |
Hb_039145_010 |
0.196062781 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 13 |
Hb_014894_010 |
0.1963101549 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 14 |
Hb_119494_010 |
0.2009641426 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 15 |
Hb_000879_190 |
0.2024297909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000853_060 |
0.2067303742 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003207_010 |
0.2075030223 |
- |
- |
PREDICTED: phospholipase D beta 1-like [Jatropha curcas] |
| 18 |
Hb_006453_010 |
0.2079436266 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 19 |
Hb_002005_100 |
0.2115531471 |
- |
- |
- |
| 20 |
Hb_012545_090 |
0.2128917776 |
- |
- |
hypothetical protein JCGZ_04320 [Jatropha curcas] |