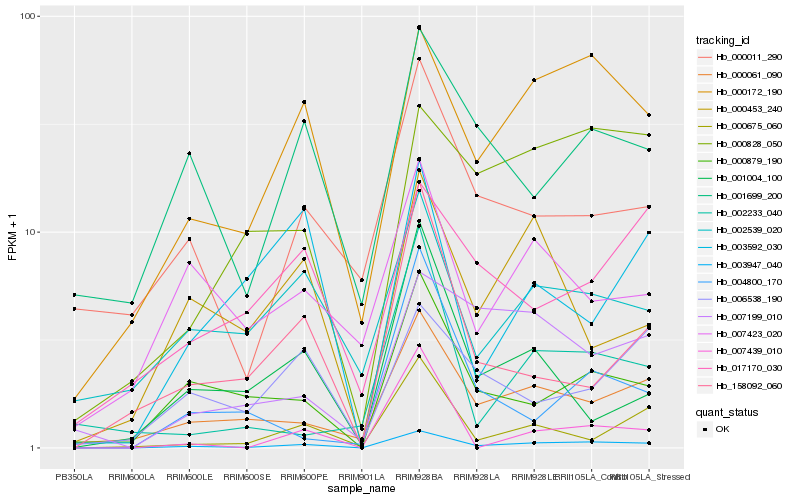
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003947_040 |
0.0 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 2 |
Hb_000172_190 |
0.1821170952 |
- |
- |
PREDICTED: uncharacterized protein LOC105650824 [Jatropha curcas] |
| 3 |
Hb_000061_090 |
0.1941459102 |
- |
- |
hypothetical protein B456_003G0981001, partial [Gossypium raimondii] |
| 4 |
Hb_000879_190 |
0.210288507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000675_060 |
0.2302394017 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_000828_050 |
0.2386394332 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_001699_200 |
0.2414448801 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002539_020 |
0.242468083 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 9 |
Hb_000011_290 |
0.2439960877 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 10 |
Hb_007199_010 |
0.2515009939 |
- |
- |
PREDICTED: protein kinase APK1A, chloroplastic [Nelumbo nucifera] |
| 11 |
Hb_158092_060 |
0.2544916217 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_007423_020 |
0.2555559624 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 13 |
Hb_000453_240 |
0.2562397944 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 14 |
Hb_006538_190 |
0.2599781396 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007439_010 |
0.2616593109 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 16 |
Hb_001004_100 |
0.265300109 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_002233_040 |
0.2718471126 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 18 |
Hb_004800_170 |
0.2730914072 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 19 |
Hb_017170_030 |
0.2733056566 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 20 |
Hb_003592_030 |
0.2740468584 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |