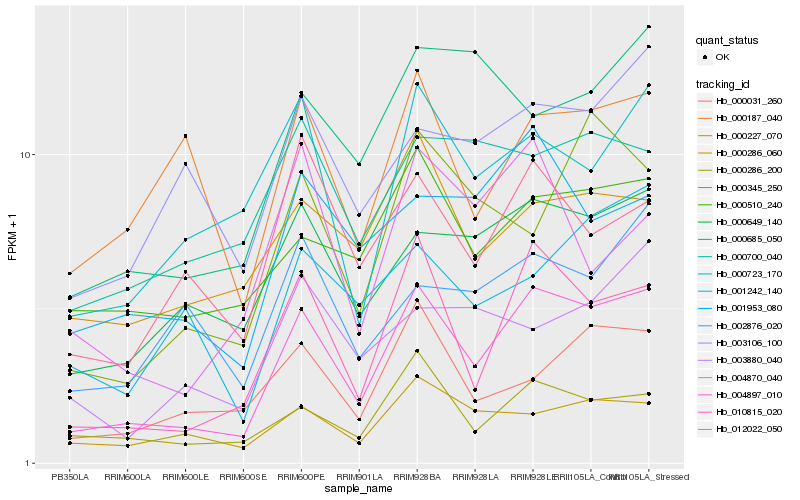
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004897_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_010815_020 |
0.1103038022 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_000286_200 |
0.1436920922 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000649_140 |
0.1515215644 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase setd3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000031_260 |
0.1594983691 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 6 |
Hb_000286_060 |
0.1604390108 |
- |
- |
PREDICTED: uncharacterized protein LOC105633357 [Jatropha curcas] |
| 7 |
Hb_001953_080 |
0.1660829837 |
- |
- |
pseudouridine synthase family protein, partial [Manihot esculenta] |
| 8 |
Hb_000510_240 |
0.1661259115 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000345_250 |
0.1671498093 |
- |
- |
cullin 1-like protein [Hevea brasiliensis] |
| 10 |
Hb_012022_050 |
0.1680876381 |
- |
- |
hypothetical protein EUGRSUZ_B02804 [Eucalyptus grandis] |
| 11 |
Hb_000723_170 |
0.168703441 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 12 |
Hb_000685_050 |
0.1708880377 |
- |
- |
PREDICTED: mitochondrial inner membrane protein OXA1-like [Populus euphratica] |
| 13 |
Hb_003106_100 |
0.1744105172 |
- |
- |
mannose-1-phosphate guanyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000227_070 |
0.1765807587 |
- |
- |
PREDICTED: ATP-dependent (S)-NAD(P)H-hydrate dehydratase isoform X2 [Jatropha curcas] |
| 15 |
Hb_001242_140 |
0.1773013436 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 16 |
Hb_004870_040 |
0.1804931405 |
- |
- |
- |
| 17 |
Hb_002876_020 |
0.1823931239 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000187_040 |
0.1824604832 |
- |
- |
PREDICTED: pyridoxal kinase isoform X5 [Vitis vinifera] |
| 19 |
Hb_003880_040 |
0.1833889874 |
- |
- |
hypothetical protein POPTR_0001s30770g [Populus trichocarpa] |
| 20 |
Hb_000700_040 |
0.1839033745 |
- |
- |
UDP-Glycosyltransferase superfamily protein isoform 4 [Theobroma cacao] |