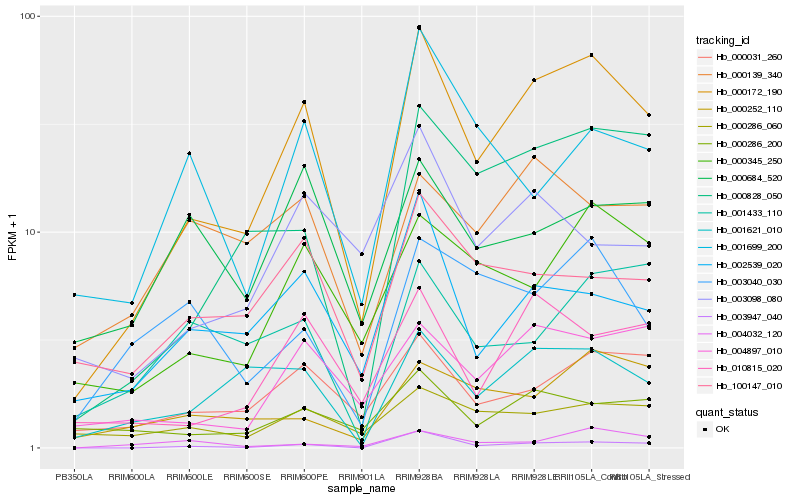
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650824 [Jatropha curcas] |
| 2 |
Hb_000828_050 |
0.1692959165 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_010815_020 |
0.1793337281 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000031_260 |
0.1796598712 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 5 |
Hb_003947_040 |
0.1821170952 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 6 |
Hb_000345_250 |
0.182284318 |
- |
- |
cullin 1-like protein [Hevea brasiliensis] |
| 7 |
Hb_004897_010 |
0.1845920309 |
- |
- |
- |
| 8 |
Hb_001621_010 |
0.1861427595 |
- |
- |
PREDICTED: uncharacterized protein LOC102615432 [Citrus sinensis] |
| 9 |
Hb_002539_020 |
0.1965883396 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 10 |
Hb_000286_200 |
0.209400986 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000286_060 |
0.2111626831 |
- |
- |
PREDICTED: uncharacterized protein LOC105633357 [Jatropha curcas] |
| 12 |
Hb_100147_010 |
0.21167902 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 13 |
Hb_003098_080 |
0.2206229447 |
- |
- |
- |
| 14 |
Hb_000252_110 |
0.2206486765 |
- |
- |
- |
| 15 |
Hb_000139_340 |
0.2260835273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004032_120 |
0.2275491885 |
- |
- |
PREDICTED: uncharacterized protein LOC104896902 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_003040_030 |
0.22780366 |
- |
- |
PREDICTED: NADP-dependent malic enzyme isoform X1 [Jatropha curcas] |
| 18 |
Hb_001699_200 |
0.2299302892 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000684_520 |
0.2301850398 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 20 |
Hb_001433_110 |
0.2312587424 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |