| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
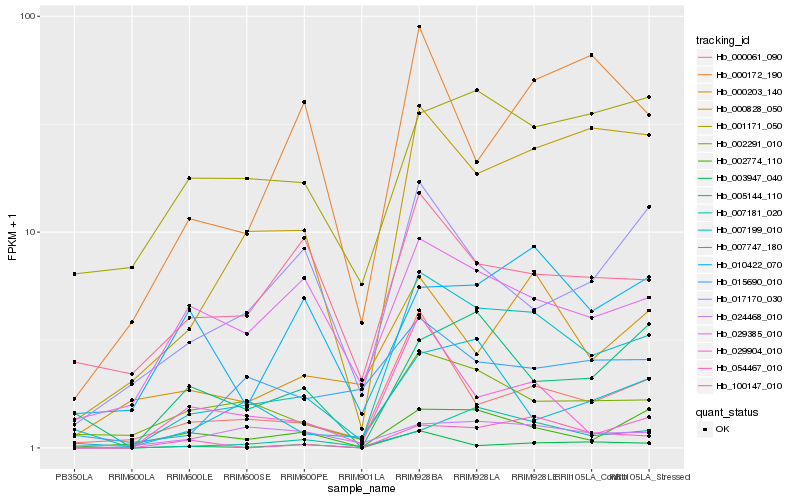
Hb_007199_010 |
0.0 |
- |
- |
PREDICTED: protein kinase APK1A, chloroplastic [Nelumbo nucifera] |
| 2 |
Hb_000828_050 |
0.1900900705 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_000061_090 |
0.2017811015 |
- |
- |
hypothetical protein B456_003G0981001, partial [Gossypium raimondii] |
| 4 |
Hb_002291_010 |
0.2149130817 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 5 |
Hb_007181_020 |
0.22197766 |
- |
- |
PREDICTED: uncharacterized protein At4g04980-like [Populus euphratica] |
| 6 |
Hb_054467_010 |
0.2306522921 |
- |
- |
- |
| 7 |
Hb_002774_110 |
0.2372433174 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_015690_010 |
0.2396298406 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CISIN_1g038521mg [Citrus sinensis] |
| 9 |
Hb_005144_110 |
0.240755841 |
- |
- |
PREDICTED: uncharacterized protein LOC104455487 [Eucalyptus grandis] |
| 10 |
Hb_000203_140 |
0.2410555244 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 11 |
Hb_007747_180 |
0.2431418901 |
transcription factor |
TF Family: TCP |
PREDICTED: bifunctional nitrilase/nitrile hydratase NIT4B-like [Jatropha curcas] |
| 12 |
Hb_001171_050 |
0.2510020745 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0284459 [Jatropha curcas] |
| 13 |
Hb_003947_040 |
0.2515009939 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 14 |
Hb_000172_190 |
0.2543359415 |
- |
- |
PREDICTED: uncharacterized protein LOC105650824 [Jatropha curcas] |
| 15 |
Hb_029385_010 |
0.260633335 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |
| 16 |
Hb_100147_010 |
0.2621364637 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 17 |
Hb_029904_010 |
0.2638098504 |
- |
- |
PREDICTED: uncharacterized protein LOC105635075 [Jatropha curcas] |
| 18 |
Hb_017170_030 |
0.2659230031 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 19 |
Hb_024468_010 |
0.2674916937 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 20 |
Hb_010422_070 |
0.2677991378 |
- |
- |
PREDICTED: UPF0483 protein CBG03338-like [Jatropha curcas] |