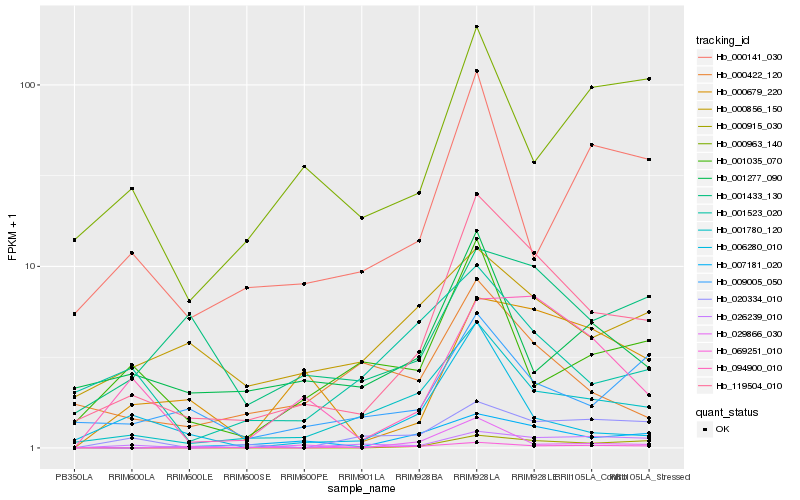
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_119504_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_15304 [Jatropha curcas] |
| 2 |
Hb_001780_120 |
0.1873711569 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 3 |
Hb_007181_020 |
0.2227912544 |
- |
- |
PREDICTED: uncharacterized protein At4g04980-like [Populus euphratica] |
| 4 |
Hb_094900_010 |
0.2426323801 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_009005_050 |
0.2538795975 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 6 |
Hb_000915_030 |
0.2542575796 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 7 |
Hb_000679_220 |
0.2634994133 |
- |
- |
PREDICTED: uncharacterized protein LOC105637638 [Jatropha curcas] |
| 8 |
Hb_001433_130 |
0.2665343342 |
- |
- |
PREDICTED: uncharacterized protein LOC105647468 isoform X1 [Jatropha curcas] |
| 9 |
Hb_026239_010 |
0.2676590679 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 10 |
Hb_020334_010 |
0.2716327294 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 11 |
Hb_000422_120 |
0.2748773097 |
- |
- |
PREDICTED: 1-phosphatidylinositol-3-phosphate 5-kinase FAB1B [Jatropha curcas] |
| 12 |
Hb_001523_020 |
0.2761567254 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 13 |
Hb_069251_010 |
0.2803695739 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_001277_090 |
0.2807458639 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 15 |
Hb_000141_030 |
0.2822101453 |
- |
- |
PREDICTED: scarecrow-like protein 22 [Jatropha curcas] |
| 16 |
Hb_001035_070 |
0.2857577142 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.7 [Jatropha curcas] |
| 17 |
Hb_029866_030 |
0.2878582209 |
- |
- |
hypothetical protein CICLE_v10006898mg [Citrus clementina] |
| 18 |
Hb_000856_150 |
0.2882053986 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 19 |
Hb_006280_010 |
0.2894815273 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor TOE3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000963_140 |
0.29145668 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |