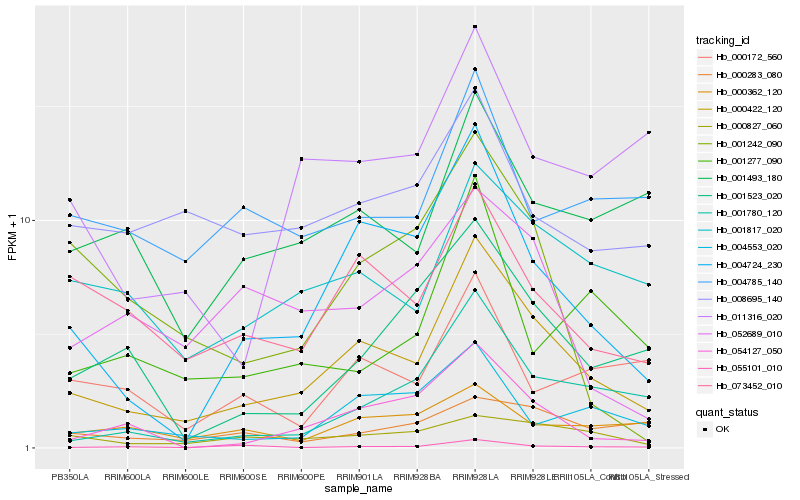
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000422_120 |
0.0 |
- |
- |
PREDICTED: 1-phosphatidylinositol-3-phosphate 5-kinase FAB1B [Jatropha curcas] |
| 2 |
Hb_004724_230 |
0.1503197367 |
- |
- |
hypothetical protein JCGZ_12701 [Jatropha curcas] |
| 3 |
Hb_001817_020 |
0.1912031644 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 4 |
Hb_073452_010 |
0.1931357295 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 5 |
Hb_054127_050 |
0.1975424468 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001780_120 |
0.2034506544 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 7 |
Hb_000827_060 |
0.2041390128 |
- |
- |
hypothetical protein POPTR_2151s00200g [Populus trichocarpa] |
| 8 |
Hb_001523_020 |
0.2090503391 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 9 |
Hb_004553_020 |
0.2095353713 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 1 homolog [Jatropha curcas] |
| 10 |
Hb_055101_010 |
0.2100436458 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_001493_180 |
0.2139232008 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 12 |
Hb_011316_020 |
0.2172531261 |
- |
- |
PREDICTED: uncharacterized protein LOC105648558 [Jatropha curcas] |
| 13 |
Hb_052689_010 |
0.2271059774 |
- |
- |
heat shock cognate protein [Solanum commersonii] |
| 14 |
Hb_001242_090 |
0.2282722115 |
- |
- |
40S ribosomal protein S18 [Colletotrichum fioriniae PJ7] |
| 15 |
Hb_001277_090 |
0.2289490537 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 16 |
Hb_000362_120 |
0.2318250065 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 17 |
Hb_008695_140 |
0.2321518996 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 18 |
Hb_004785_140 |
0.2326700466 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000172_560 |
0.2339296255 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750 [Jatropha curcas] |
| 20 |
Hb_000283_080 |
0.235991763 |
- |
- |
- |