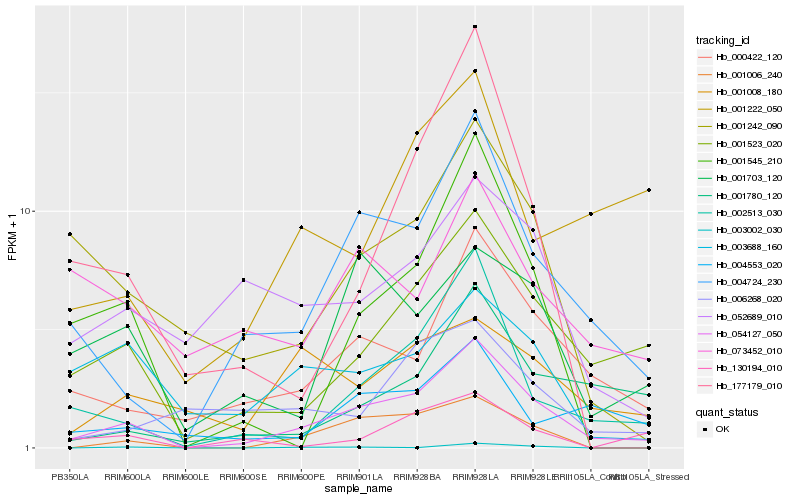
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054127_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004724_230 |
0.1788323729 |
- |
- |
hypothetical protein JCGZ_12701 [Jatropha curcas] |
| 3 |
Hb_001523_020 |
0.1863016056 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 4 |
Hb_000422_120 |
0.1975424468 |
- |
- |
PREDICTED: 1-phosphatidylinositol-3-phosphate 5-kinase FAB1B [Jatropha curcas] |
| 5 |
Hb_002513_030 |
0.2056996116 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X2 [Elaeis guineensis] |
| 6 |
Hb_001006_240 |
0.2230576752 |
- |
- |
- |
| 7 |
Hb_001242_090 |
0.2398186689 |
- |
- |
40S ribosomal protein S18 [Colletotrichum fioriniae PJ7] |
| 8 |
Hb_001545_210 |
0.2481119502 |
- |
- |
hypothetical protein JCGZ_11132 [Jatropha curcas] |
| 9 |
Hb_001780_120 |
0.2481215055 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 10 |
Hb_004553_020 |
0.2488140066 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 1 homolog [Jatropha curcas] |
| 11 |
Hb_001703_120 |
0.2546367883 |
- |
- |
PREDICTED: uncharacterized protein LOC105116966 isoform X2 [Populus euphratica] |
| 12 |
Hb_003688_160 |
0.2595790264 |
- |
- |
Presenilin-like family protein [Populus trichocarpa] |
| 13 |
Hb_052689_010 |
0.2615767217 |
- |
- |
heat shock cognate protein [Solanum commersonii] |
| 14 |
Hb_001008_180 |
0.2646368051 |
- |
- |
- |
| 15 |
Hb_001222_050 |
0.2663380757 |
- |
- |
hypothetical protein CICLE_v100161911mg, partial [Citrus clementina] |
| 16 |
Hb_177179_010 |
0.2665252244 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Populus euphratica] |
| 17 |
Hb_130194_010 |
0.2665560591 |
- |
- |
PREDICTED: uncharacterized protein LOC104115665 [Nicotiana tomentosiformis] |
| 18 |
Hb_006268_020 |
0.268191351 |
- |
- |
hypothetical protein JCGZ_10516 [Jatropha curcas] |
| 19 |
Hb_003002_030 |
0.2755343593 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 20 |
Hb_073452_010 |
0.2776001322 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |