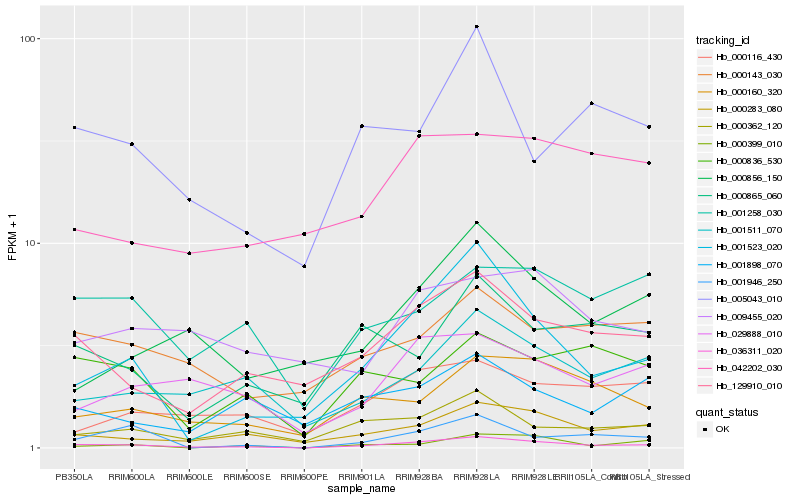
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_036311_020 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_001523_020 |
0.157070922 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 3 |
Hb_000283_080 |
0.1755284476 |
- |
- |
- |
| 4 |
Hb_129910_010 |
0.1875856951 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1X [Populus euphratica] |
| 5 |
Hb_000362_120 |
0.2008904049 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 6 |
Hb_001511_070 |
0.2016704239 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 7 |
Hb_000160_320 |
0.2028296053 |
- |
- |
hypothetical protein POPTR_0004s08730g [Populus trichocarpa] |
| 8 |
Hb_001946_250 |
0.2101106868 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 18 [Orycteropus afer afer] |
| 9 |
Hb_005043_010 |
0.2120416846 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_029888_010 |
0.2142361968 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Nicotiana tomentosiformis] |
| 11 |
Hb_000865_060 |
0.2222084754 |
transcription factor |
TF Family: FAR1 |
hypothetical protein CISIN_1g043648mg [Citrus sinensis] |
| 12 |
Hb_000143_030 |
0.2231860512 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 13 |
Hb_000856_150 |
0.2243872125 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 14 |
Hb_000399_010 |
0.2248838554 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 15 |
Hb_001258_030 |
0.2251220246 |
- |
- |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 16 |
Hb_000116_430 |
0.226951073 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000836_530 |
0.2273571353 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 18 |
Hb_001898_070 |
0.2355095887 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16610 [Jatropha curcas] |
| 19 |
Hb_009455_020 |
0.2360460348 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 20 |
Hb_042202_030 |
0.2410252048 |
- |
- |
PREDICTED: uncharacterized protein LOC105647255 [Jatropha curcas] |