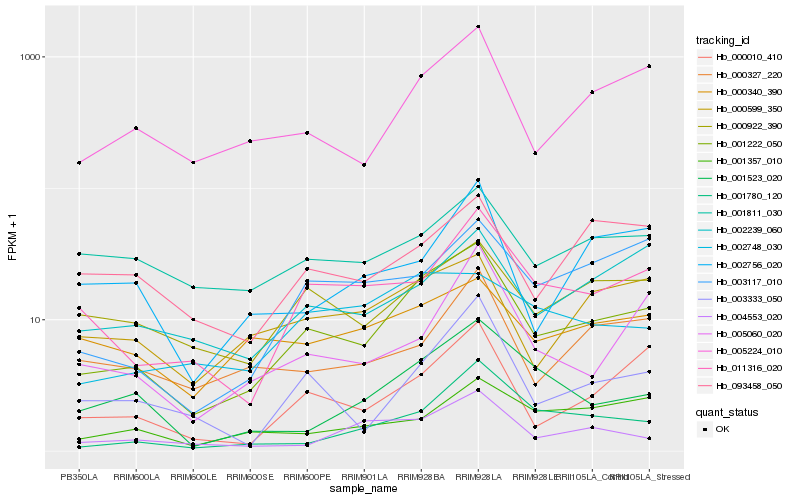
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001222_050 |
0.0 |
- |
- |
hypothetical protein CICLE_v100161911mg, partial [Citrus clementina] |
| 2 |
Hb_005224_010 |
0.1607006939 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 3 |
Hb_011316_020 |
0.1683868139 |
- |
- |
PREDICTED: uncharacterized protein LOC105648558 [Jatropha curcas] |
| 4 |
Hb_000922_390 |
0.1760131086 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 5 |
Hb_000010_410 |
0.1768612492 |
- |
- |
hypothetical protein POPTR_0015s10070g, partial [Populus trichocarpa] |
| 6 |
Hb_003333_050 |
0.1815109622 |
- |
- |
hypothetical protein JCGZ_20906 [Jatropha curcas] |
| 7 |
Hb_000599_350 |
0.1870187875 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 8 |
Hb_002239_060 |
0.1885390498 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 9 |
Hb_003117_010 |
0.1890533608 |
- |
- |
PREDICTED: uncharacterized protein LOC100259103 isoform X1 [Vitis vinifera] |
| 10 |
Hb_001523_020 |
0.1947634558 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 11 |
Hb_001780_120 |
0.1956387547 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 12 |
Hb_004553_020 |
0.200965575 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 1 homolog [Jatropha curcas] |
| 13 |
Hb_005060_020 |
0.2011892236 |
- |
- |
PREDICTED: metal tolerance protein 10-like isoform X1 [Populus euphratica] |
| 14 |
Hb_002748_030 |
0.2027577465 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 15 |
Hb_000327_220 |
0.2031573884 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 16 |
Hb_000340_390 |
0.2034390362 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 17 |
Hb_093458_050 |
0.2045049265 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 18 |
Hb_001357_010 |
0.2053110101 |
- |
- |
- |
| 19 |
Hb_001811_030 |
0.2059289942 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 20 |
Hb_002756_020 |
0.2066133404 |
transcription factor |
TF Family: HB |
bel1 homeotic protein, putative [Ricinus communis] |