| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
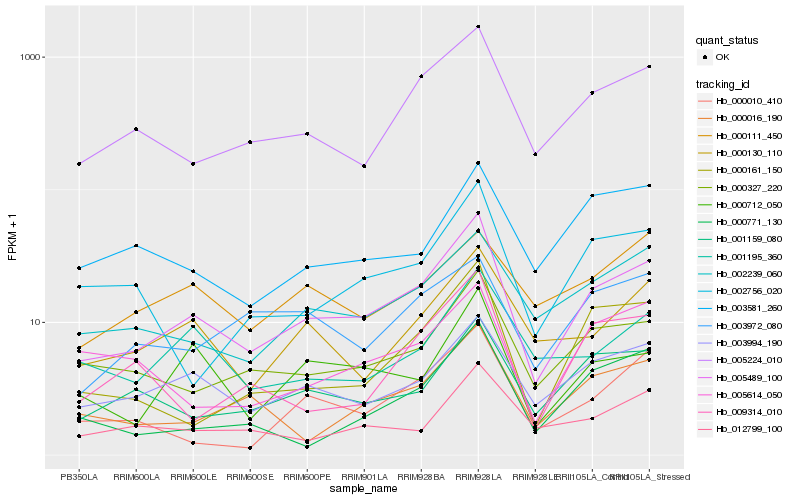
Hb_005489_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003994_190 |
0.1330007068 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005224_010 |
0.1334255292 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 4 |
Hb_000327_220 |
0.1386241155 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 5 |
Hb_001159_080 |
0.1500057856 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |
| 6 |
Hb_002239_060 |
0.1522359529 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 7 |
Hb_000130_110 |
0.1593862372 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 8 |
Hb_000712_050 |
0.1623955691 |
- |
- |
PREDICTED: inositol oxygenase 1-like isoform X1 [Citrus sinensis] |
| 9 |
Hb_000016_190 |
0.1642509122 |
- |
- |
ABC transporter B family member 28 [Glycine soja] |
| 10 |
Hb_000161_150 |
0.1675896126 |
- |
- |
PREDICTED: protein argonaute 2-like [Jatropha curcas] |
| 11 |
Hb_000010_410 |
0.1708517187 |
- |
- |
hypothetical protein POPTR_0015s10070g, partial [Populus trichocarpa] |
| 12 |
Hb_012799_100 |
0.1722198708 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 13 |
Hb_002756_020 |
0.1740896254 |
transcription factor |
TF Family: HB |
bel1 homeotic protein, putative [Ricinus communis] |
| 14 |
Hb_001195_360 |
0.1753110068 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 15 |
Hb_003581_260 |
0.1764225757 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29-like [Jatropha curcas] |
| 16 |
Hb_000771_130 |
0.1786468901 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 17 |
Hb_003972_080 |
0.1795560502 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 18 |
Hb_005614_050 |
0.1834791593 |
- |
- |
PREDICTED: aspartate aminotransferase, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_009314_010 |
0.1842115982 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 20 |
Hb_000111_450 |
0.1872714812 |
- |
- |
PREDICTED: probable choline kinase 2 [Jatropha curcas] |