| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
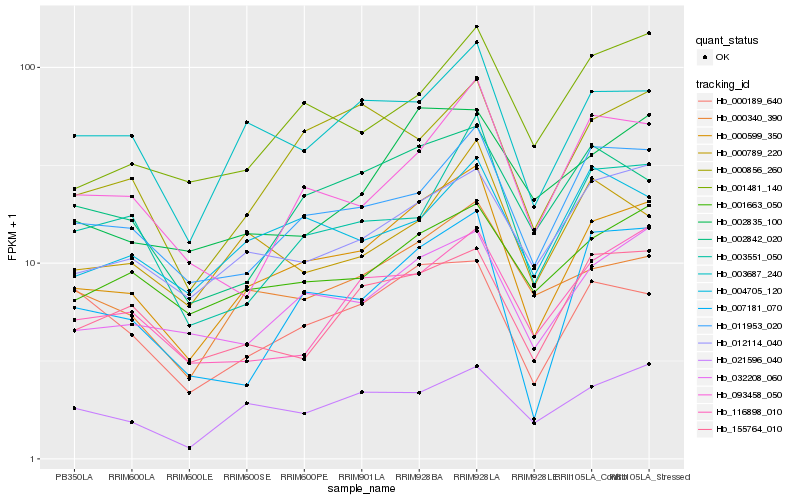
Hb_000599_350 |
0.0 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 2 |
Hb_032208_060 |
0.1074978669 |
- |
- |
PREDICTED: constitutive photomorphogenesis protein 10 isoform X2 [Populus euphratica] |
| 3 |
Hb_003687_240 |
0.1117976292 |
- |
- |
PREDICTED: cryptochrome-1 [Jatropha curcas] |
| 4 |
Hb_011953_020 |
0.1136862876 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 5 |
Hb_000340_390 |
0.1164942977 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 6 |
Hb_012114_040 |
0.1196995832 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_007181_070 |
0.1228551845 |
- |
- |
PREDICTED: alpha-galactosidase-like [Jatropha curcas] |
| 8 |
Hb_021596_040 |
0.1247566939 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_001663_050 |
0.1273406475 |
- |
- |
PREDICTED: GPN-loop GTPase 3 [Jatropha curcas] |
| 10 |
Hb_000856_260 |
0.1296862231 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0015s07640g [Populus trichocarpa] |
| 11 |
Hb_002835_100 |
0.1310109604 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 12 |
Hb_002842_020 |
0.1314129154 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 13 |
Hb_093458_050 |
0.1322619981 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 14 |
Hb_001481_140 |
0.1342528586 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 15 |
Hb_155764_010 |
0.1356028984 |
- |
- |
- |
| 16 |
Hb_000189_640 |
0.1357265823 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 17 |
Hb_003551_050 |
0.1357545547 |
- |
- |
PREDICTED: mitochondrial fission 1 protein A [Jatropha curcas] |
| 18 |
Hb_116898_010 |
0.1375947123 |
- |
- |
PREDICTED: uncharacterized protein LOC105639569 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004705_120 |
0.1399653962 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_000789_220 |
0.1405038724 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |