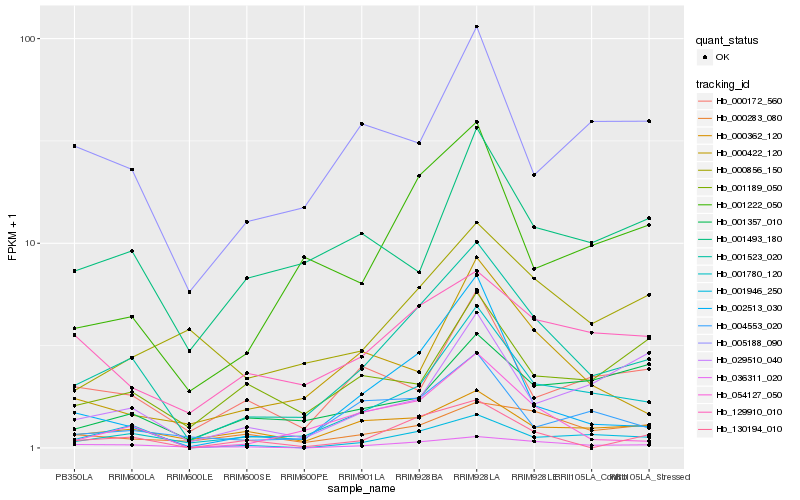
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001523_020 |
0.0 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 2 |
Hb_036311_020 |
0.157070922 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_001780_120 |
0.1771888034 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 4 |
Hb_054127_050 |
0.1863016056 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001222_050 |
0.1947634558 |
- |
- |
hypothetical protein CICLE_v100161911mg, partial [Citrus clementina] |
| 6 |
Hb_000362_120 |
0.198553908 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 7 |
Hb_130194_010 |
0.2006466559 |
- |
- |
PREDICTED: uncharacterized protein LOC104115665 [Nicotiana tomentosiformis] |
| 8 |
Hb_000422_120 |
0.2090503391 |
- |
- |
PREDICTED: 1-phosphatidylinositol-3-phosphate 5-kinase FAB1B [Jatropha curcas] |
| 9 |
Hb_129910_010 |
0.2093571647 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1X [Populus euphratica] |
| 10 |
Hb_004553_020 |
0.2103177367 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 1 homolog [Jatropha curcas] |
| 11 |
Hb_000283_080 |
0.2117195128 |
- |
- |
- |
| 12 |
Hb_002513_030 |
0.2153810595 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X2 [Elaeis guineensis] |
| 13 |
Hb_000856_150 |
0.2166011358 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 14 |
Hb_001946_250 |
0.2211381511 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 18 [Orycteropus afer afer] |
| 15 |
Hb_005188_090 |
0.2233138934 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase gamma 7-like [Jatropha curcas] |
| 16 |
Hb_001493_180 |
0.2284947223 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 17 |
Hb_000172_560 |
0.229658745 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750 [Jatropha curcas] |
| 18 |
Hb_001357_010 |
0.2298689125 |
- |
- |
- |
| 19 |
Hb_029510_040 |
0.2309315635 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Vitis vinifera] |
| 20 |
Hb_001189_050 |
0.2336286558 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |