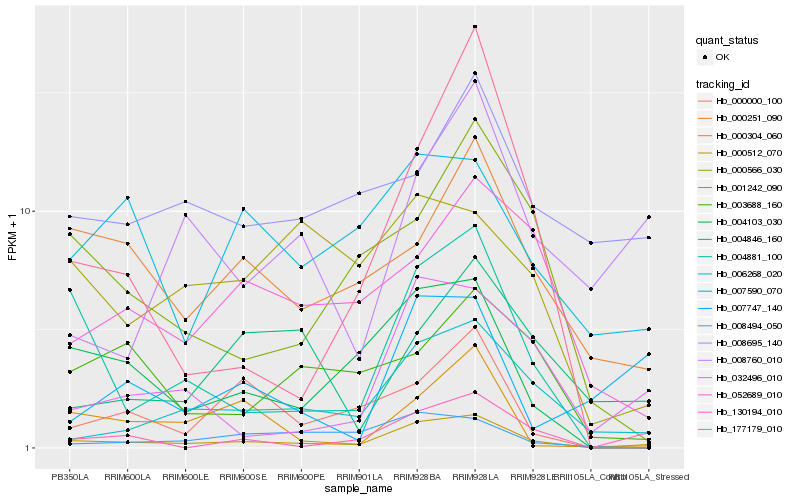
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000512_070 |
0.0 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71250-like [Jatropha curcas] |
| 2 |
Hb_006268_020 |
0.2041349997 |
- |
- |
hypothetical protein JCGZ_10516 [Jatropha curcas] |
| 3 |
Hb_004103_030 |
0.2149288207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_130194_010 |
0.218346456 |
- |
- |
PREDICTED: uncharacterized protein LOC104115665 [Nicotiana tomentosiformis] |
| 5 |
Hb_032496_010 |
0.2213033881 |
- |
- |
hypothetical protein POPTR_0012s10850g [Populus trichocarpa] |
| 6 |
Hb_004881_100 |
0.2277535809 |
- |
- |
Phosphoribosyltransferase family protein isoform 4 [Theobroma cacao] |
| 7 |
Hb_000251_090 |
0.2427595271 |
- |
- |
PREDICTED: two-pore potassium channel 3-like [Jatropha curcas] |
| 8 |
Hb_007590_070 |
0.2577450946 |
- |
- |
PREDICTED: inositol 3-kinase [Jatropha curcas] |
| 9 |
Hb_001242_090 |
0.2585904698 |
- |
- |
40S ribosomal protein S18 [Colletotrichum fioriniae PJ7] |
| 10 |
Hb_052689_010 |
0.2592695123 |
- |
- |
heat shock cognate protein [Solanum commersonii] |
| 11 |
Hb_000000_100 |
0.2626395145 |
- |
- |
hypothetical protein POPTR_0002s13260g [Populus trichocarpa] |
| 12 |
Hb_008695_140 |
0.2671475813 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 13 |
Hb_000304_060 |
0.2693788093 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0010s15360g [Populus trichocarpa] |
| 14 |
Hb_008494_050 |
0.2725098559 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 15 |
Hb_008760_010 |
0.274044718 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 16 |
Hb_000566_030 |
0.276843442 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 17 |
Hb_007747_140 |
0.2770111183 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_177179_010 |
0.2779501227 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Populus euphratica] |
| 19 |
Hb_004846_160 |
0.2794929428 |
- |
- |
- |
| 20 |
Hb_003688_160 |
0.2802825504 |
- |
- |
Presenilin-like family protein [Populus trichocarpa] |