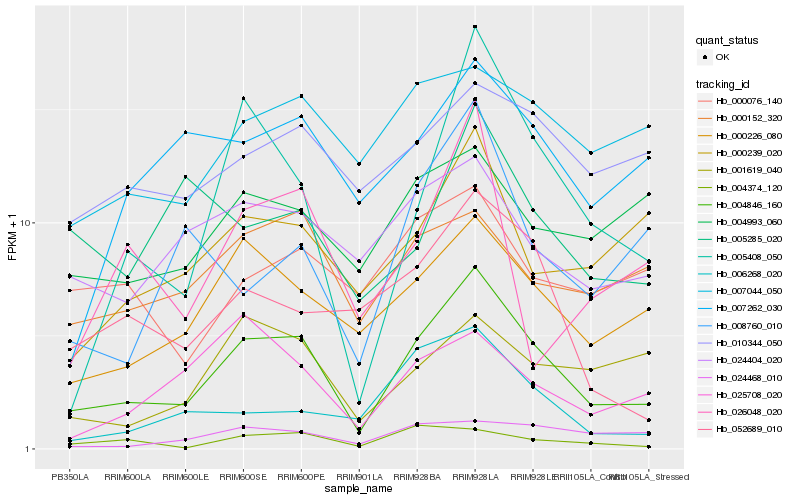
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004846_160 |
0.0 |
- |
- |
- |
| 2 |
Hb_000226_080 |
0.1756515545 |
- |
- |
prephenate dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_005408_050 |
0.1885803024 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK5 [Jatropha curcas] |
| 4 |
Hb_000239_020 |
0.2048824398 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 5 |
Hb_052689_010 |
0.2065472393 |
- |
- |
heat shock cognate protein [Solanum commersonii] |
| 6 |
Hb_024404_020 |
0.2069747185 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 7 |
Hb_004374_120 |
0.2136953366 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 8 |
Hb_007262_030 |
0.2175028513 |
- |
- |
- |
| 9 |
Hb_001619_040 |
0.2180075625 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 10 |
Hb_000152_320 |
0.2180822587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006268_020 |
0.2184186456 |
- |
- |
hypothetical protein JCGZ_10516 [Jatropha curcas] |
| 12 |
Hb_007044_050 |
0.2190954137 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |
| 13 |
Hb_010344_050 |
0.2231321621 |
- |
- |
Adagio 1 -like protein [Gossypium arboreum] |
| 14 |
Hb_004993_060 |
0.2232772804 |
- |
- |
- |
| 15 |
Hb_025708_020 |
0.2237252004 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 16 |
Hb_008760_010 |
0.2244465622 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 17 |
Hb_024468_010 |
0.2256456101 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 18 |
Hb_005285_020 |
0.2273728484 |
- |
- |
- |
| 19 |
Hb_000076_140 |
0.2278422908 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 1, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_026048_020 |
0.2294313688 |
- |
- |
PREDICTED: uncharacterized protein LOC105645738 [Jatropha curcas] |