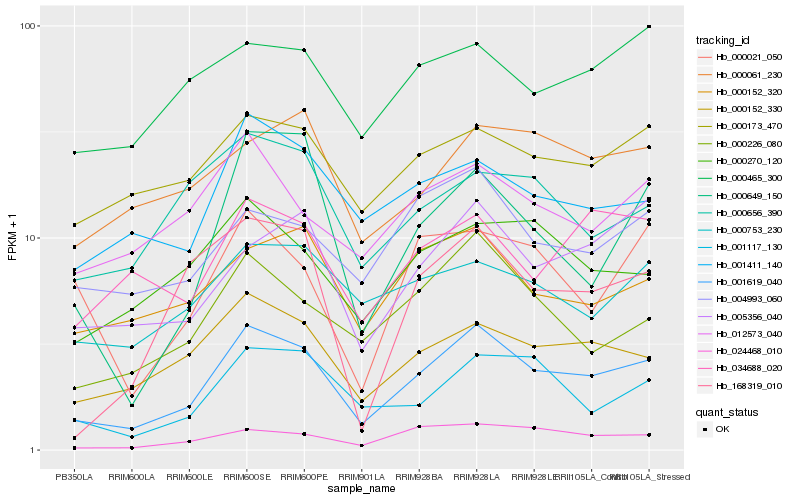
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001619_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 2 |
Hb_000152_330 |
0.1301612287 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 3 |
Hb_000226_080 |
0.1379644024 |
- |
- |
prephenate dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_024468_010 |
0.1438760789 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 5 |
Hb_001117_130 |
0.1454189045 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |
| 6 |
Hb_005356_040 |
0.1475714056 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 7 |
Hb_000649_150 |
0.1509583738 |
- |
- |
PREDICTED: uncharacterized protein LOC105634115 [Jatropha curcas] |
| 8 |
Hb_001411_140 |
0.152542396 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 9 |
Hb_004993_060 |
0.1537529707 |
- |
- |
- |
| 10 |
Hb_000152_320 |
0.1547746016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000270_120 |
0.1566664304 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 12 |
Hb_000173_470 |
0.158387106 |
- |
- |
PREDICTED: BSD domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_012573_040 |
0.1596328022 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g55270-like [Jatropha curcas] |
| 14 |
Hb_034688_020 |
0.1615274936 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 15 |
Hb_000753_230 |
0.161735086 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000061_230 |
0.1646665397 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 17 |
Hb_000021_050 |
0.1653255567 |
- |
- |
PREDICTED: uncharacterized protein LOC105645238 [Jatropha curcas] |
| 18 |
Hb_168319_010 |
0.1655638826 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000465_300 |
0.1658805711 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 20 |
Hb_000656_390 |
0.1660395984 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |