| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
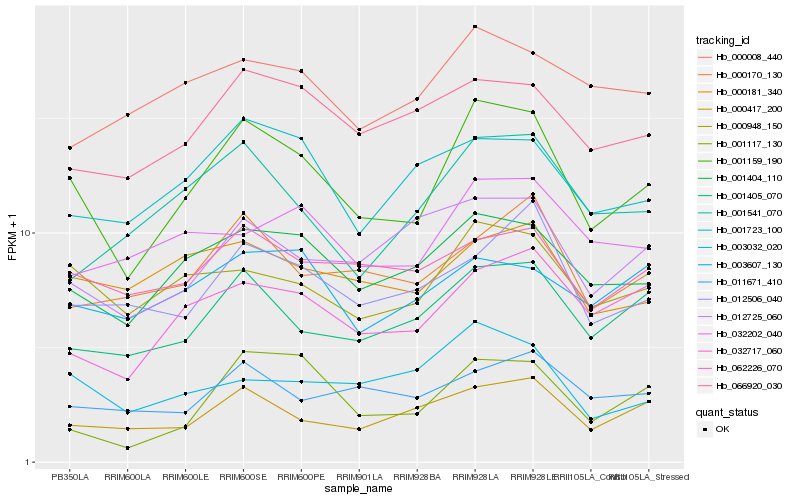
Hb_001159_190 |
0.0 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_001404_110 |
0.1075796425 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 3 |
Hb_000948_150 |
0.1163382319 |
- |
- |
PREDICTED: heme-binding-like protein At3g10130, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001405_070 |
0.1201710199 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 5 |
Hb_032717_060 |
0.1251609719 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ18 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000417_200 |
0.1287012416 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_066920_030 |
0.132948201 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 8 |
Hb_001723_100 |
0.1347381055 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000170_130 |
0.1357853752 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 10 |
Hb_062226_070 |
0.1359432584 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_012506_040 |
0.136178748 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 12 |
Hb_012725_060 |
0.1394498308 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_000181_340 |
0.1403172778 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 14 |
Hb_003032_020 |
0.1408746396 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55740, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001117_130 |
0.1425072737 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |
| 16 |
Hb_001541_070 |
0.1432240102 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1-like [Jatropha curcas] |
| 17 |
Hb_011671_410 |
0.1432556456 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 18 |
Hb_032202_040 |
0.1449823743 |
- |
- |
PREDICTED: phospholipase D beta 2 [Jatropha curcas] |
| 19 |
Hb_003607_130 |
0.1475959188 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000008_440 |
0.1488666908 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LOG2 [Jatropha curcas] |