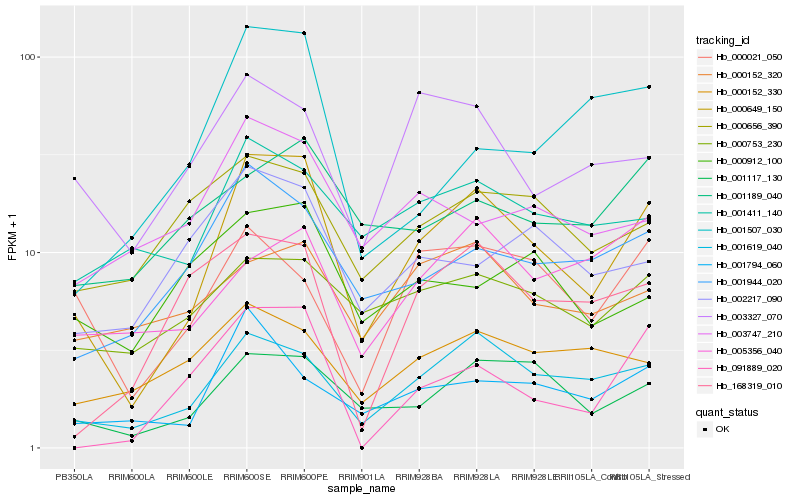
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634115 [Jatropha curcas] |
| 2 |
Hb_001619_040 |
0.1509583738 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 3 |
Hb_001117_130 |
0.1625668583 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |
| 4 |
Hb_001944_020 |
0.1926390092 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 5 |
Hb_001794_060 |
0.2047200404 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001411_140 |
0.2111029816 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 7 |
Hb_000753_230 |
0.2118450765 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000021_050 |
0.2119009452 |
- |
- |
PREDICTED: uncharacterized protein LOC105645238 [Jatropha curcas] |
| 9 |
Hb_003327_070 |
0.2148789605 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 10 |
Hb_091889_020 |
0.2159335414 |
- |
- |
- |
| 11 |
Hb_168319_010 |
0.217531894 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002217_090 |
0.2183366188 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 13 |
Hb_000656_390 |
0.2189243665 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000152_320 |
0.2202302231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003747_210 |
0.2221127113 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 16 |
Hb_000152_330 |
0.2233733307 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 17 |
Hb_005356_040 |
0.2246286615 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 18 |
Hb_001189_040 |
0.2248719861 |
- |
- |
hypothetical protein POPTR_0014s10420g [Populus trichocarpa] |
| 19 |
Hb_000912_100 |
0.2248860544 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 20 |
Hb_001507_030 |
0.2253242021 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |