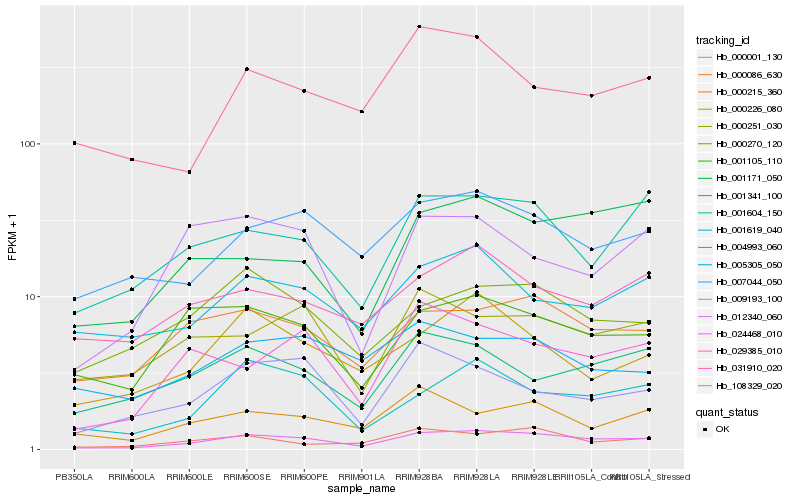
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024468_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 2 |
Hb_000086_630 |
0.1376607622 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_007044_050 |
0.1429273711 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |
| 4 |
Hb_001619_040 |
0.1438760789 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 5 |
Hb_001604_150 |
0.1461248292 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Nelumbo nucifera] |
| 6 |
Hb_001105_110 |
0.1476301272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000001_130 |
0.151902197 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 8 |
Hb_000270_120 |
0.1525314766 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 9 |
Hb_029385_010 |
0.1548680292 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |
| 10 |
Hb_000251_030 |
0.1550471258 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000226_080 |
0.1558248397 |
- |
- |
prephenate dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_001341_100 |
0.1567131376 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |
| 13 |
Hb_004993_060 |
0.157266581 |
- |
- |
- |
| 14 |
Hb_031910_020 |
0.1597066167 |
- |
- |
PREDICTED: exocyst complex component SEC15A [Jatropha curcas] |
| 15 |
Hb_009193_100 |
0.1602023861 |
- |
- |
hypothetical protein JCGZ_15709 [Jatropha curcas] |
| 16 |
Hb_001171_050 |
0.160610112 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0284459 [Jatropha curcas] |
| 17 |
Hb_108329_020 |
0.1619188272 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 18 |
Hb_000215_360 |
0.1620273075 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 19 |
Hb_012340_060 |
0.1666554492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005305_050 |
0.1709597366 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |