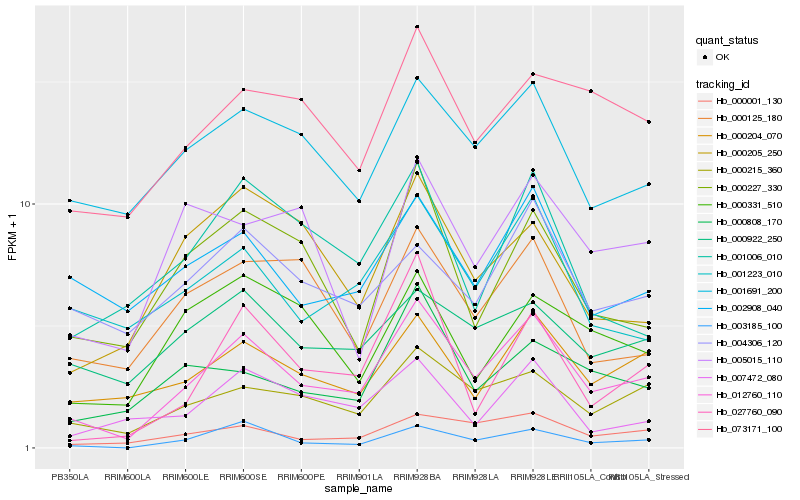
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012760_110 |
0.0 |
- |
- |
PREDICTED: methyltransferase-like protein 23 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000215_360 |
0.123981161 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 3 |
Hb_000001_130 |
0.1309892814 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 4 |
Hb_003185_100 |
0.1395319775 |
- |
- |
hypothetical protein JCGZ_07188 [Jatropha curcas] |
| 5 |
Hb_000204_070 |
0.1474555276 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 6 |
Hb_073171_100 |
0.1593322036 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 7 |
Hb_001691_200 |
0.1602993783 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 8 |
Hb_001223_010 |
0.1610298491 |
- |
- |
PREDICTED: FGGY carbohydrate kinase domain-containing protein isoform X2 [Jatropha curcas] |
| 9 |
Hb_000227_330 |
0.1635005334 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 10 |
Hb_000808_170 |
0.1644624194 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 11 |
Hb_000125_180 |
0.1677268908 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 12 |
Hb_000922_250 |
0.1679311464 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_002908_040 |
0.1690889952 |
- |
- |
DNA gyrase subunit A, chloroplast/mitochondrial precursor, putative [Ricinus communis] |
| 14 |
Hb_027760_090 |
0.1691391378 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_007472_080 |
0.1736599431 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 16 |
Hb_001006_010 |
0.1741424923 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 17 |
Hb_000331_510 |
0.1761280184 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 18 |
Hb_000205_250 |
0.1762404201 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 19 |
Hb_004306_120 |
0.1770872485 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_005015_110 |
0.1781411334 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |