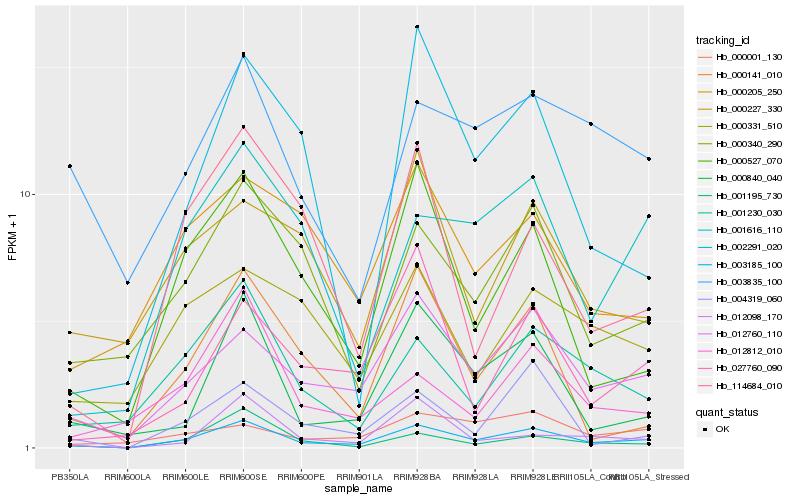
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003185_100 |
0.0 |
- |
- |
hypothetical protein JCGZ_07188 [Jatropha curcas] |
| 2 |
Hb_012760_110 |
0.1395319775 |
- |
- |
PREDICTED: methyltransferase-like protein 23 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000527_070 |
0.1776343798 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 4 |
Hb_001230_030 |
0.1817165254 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 isoform X2 [Populus euphratica] |
| 5 |
Hb_000840_040 |
0.1864158036 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 6 |
Hb_000340_290 |
0.1899955045 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog [Jatropha curcas] |
| 7 |
Hb_001616_110 |
0.1940583003 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 8 |
Hb_001195_730 |
0.1981738584 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 9 |
Hb_004319_060 |
0.2007431502 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 10 |
Hb_000141_010 |
0.2029524078 |
- |
- |
PREDICTED: uncharacterized protein LOC105632253 [Jatropha curcas] |
| 11 |
Hb_012812_010 |
0.2075433091 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 12 |
Hb_002291_020 |
0.2078331082 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_027760_090 |
0.2084613325 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_114684_010 |
0.2088417333 |
- |
- |
PREDICTED: seed maturation protein PM36 [Jatropha curcas] |
| 15 |
Hb_000001_130 |
0.2095350213 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 16 |
Hb_000331_510 |
0.2106583864 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 17 |
Hb_000205_250 |
0.2113938896 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 18 |
Hb_012098_170 |
0.2134980813 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_16874 [Jatropha curcas] |
| 19 |
Hb_000227_330 |
0.2151982635 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 20 |
Hb_003835_100 |
0.2158435555 |
- |
- |
PREDICTED: putative U-box domain-containing protein 50 [Jatropha curcas] |