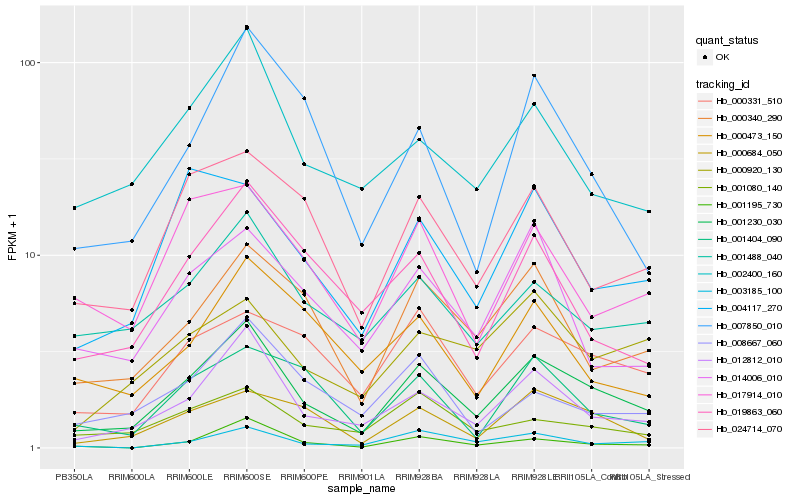
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001230_030 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 isoform X2 [Populus euphratica] |
| 2 |
Hb_012812_010 |
0.1141105962 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 3 |
Hb_000340_290 |
0.1557454937 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog [Jatropha curcas] |
| 4 |
Hb_002400_160 |
0.1596217446 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 5 |
Hb_000684_050 |
0.1597478726 |
- |
- |
PREDICTED: MLO-like protein 6 [Sesamum indicum] |
| 6 |
Hb_001488_040 |
0.162020604 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 7 |
Hb_000331_510 |
0.1629945164 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 8 |
Hb_024714_070 |
0.1650352948 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 9 |
Hb_001080_140 |
0.1693845891 |
transcription factor |
TF Family: RWP-RK |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_017914_010 |
0.171761804 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 11 |
Hb_019863_060 |
0.1744074028 |
- |
- |
PREDICTED: target of Myb protein 1-like [Populus euphratica] |
| 12 |
Hb_007850_010 |
0.1754943296 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 13 |
Hb_001195_730 |
0.1772413924 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 14 |
Hb_004117_270 |
0.1776059163 |
- |
- |
- |
| 15 |
Hb_014006_010 |
0.1807313963 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 16 |
Hb_001404_090 |
0.1808729597 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Camelina sativa] |
| 17 |
Hb_000920_130 |
0.181149514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003185_100 |
0.1817165254 |
- |
- |
hypothetical protein JCGZ_07188 [Jatropha curcas] |
| 19 |
Hb_008667_060 |
0.1824369059 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 20 |
Hb_000473_150 |
0.1828502867 |
- |
- |
PREDICTED: pullulanase 1, chloroplastic [Jatropha curcas] |