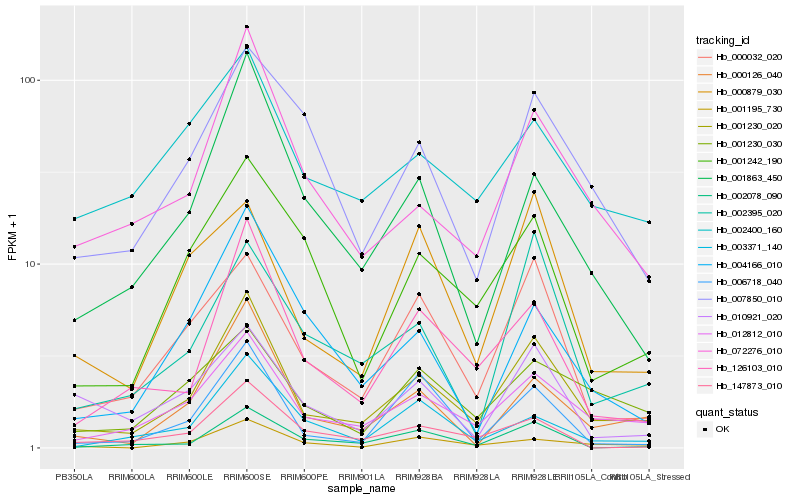
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001230_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s31610g [Populus trichocarpa] |
| 2 |
Hb_012812_010 |
0.1128455394 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 3 |
Hb_000126_040 |
0.1475528721 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_072276_010 |
0.1519399453 |
- |
- |
PREDICTED: elongation of fatty acids protein 3-like [Jatropha curcas] |
| 5 |
Hb_126103_010 |
0.1543850121 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 6 |
Hb_000032_020 |
0.1704566018 |
- |
- |
Centromeric protein E, putative [Ricinus communis] |
| 7 |
Hb_001195_730 |
0.1719642282 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 8 |
Hb_001863_450 |
0.1745053049 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 9 |
Hb_002078_090 |
0.1857296262 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_001242_190 |
0.1857884425 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 11 |
Hb_001230_030 |
0.1872606801 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 isoform X2 [Populus euphratica] |
| 12 |
Hb_002395_020 |
0.1875287362 |
- |
- |
Protein CREG1 precursor, putative [Ricinus communis] |
| 13 |
Hb_006718_040 |
0.1902900266 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_007850_010 |
0.1906788244 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 15 |
Hb_147873_010 |
0.1941232589 |
- |
- |
PREDICTED: uncharacterized protein LOC102595311 [Solanum tuberosum] |
| 16 |
Hb_010921_020 |
0.1946118395 |
- |
- |
PREDICTED: cyclin-SDS [Jatropha curcas] |
| 17 |
Hb_003371_140 |
0.1951273969 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 18 |
Hb_000879_030 |
0.1962759163 |
- |
- |
PREDICTED: uncharacterized protein LOC103707552 [Phoenix dactylifera] |
| 19 |
Hb_004166_010 |
0.1974462838 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 20 |
Hb_002400_160 |
0.1975534291 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |