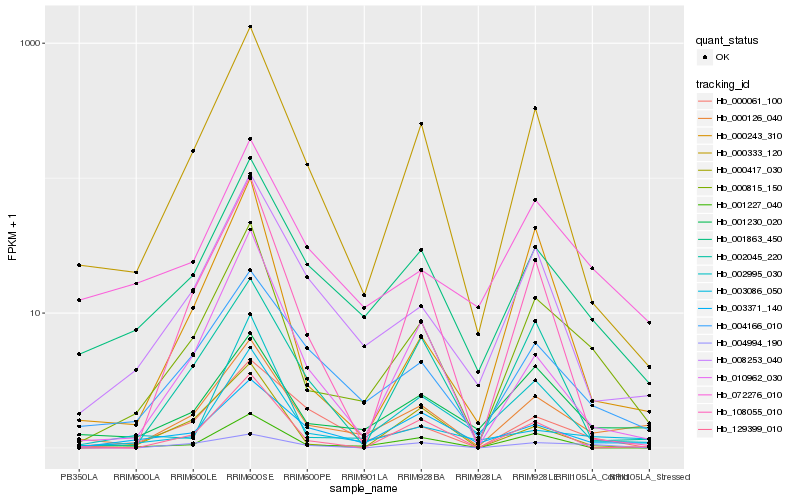
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000815_150 |
0.0 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000126_040 |
0.1635790685 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_000333_120 |
0.1747203728 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 4 |
Hb_001863_450 |
0.1792963015 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 5 |
Hb_004166_010 |
0.1997562216 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 6 |
Hb_002995_030 |
0.2004536778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001230_020 |
0.2013481511 |
- |
- |
hypothetical protein POPTR_0001s31610g [Populus trichocarpa] |
| 8 |
Hb_010962_030 |
0.2048602554 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_001227_040 |
0.204938546 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 10 |
Hb_008253_040 |
0.206877916 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 11 |
Hb_108055_010 |
0.2091293068 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000243_310 |
0.2113464108 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 13 |
Hb_003086_050 |
0.2159969428 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 14 |
Hb_129399_010 |
0.2194851787 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_004994_190 |
0.2201947673 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like [Jatropha curcas] |
| 16 |
Hb_000417_030 |
0.2203272633 |
transcription factor |
TF Family: G2-like |
PREDICTED: putative two-component response regulator ARR19 [Jatropha curcas] |
| 17 |
Hb_002045_220 |
0.2246970607 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 18 |
Hb_072276_010 |
0.2252688013 |
- |
- |
PREDICTED: elongation of fatty acids protein 3-like [Jatropha curcas] |
| 19 |
Hb_003371_140 |
0.225765777 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 20 |
Hb_000061_100 |
0.2266456554 |
- |
- |
hypothetical protein CISIN_1g0053742mg, partial [Citrus sinensis] |