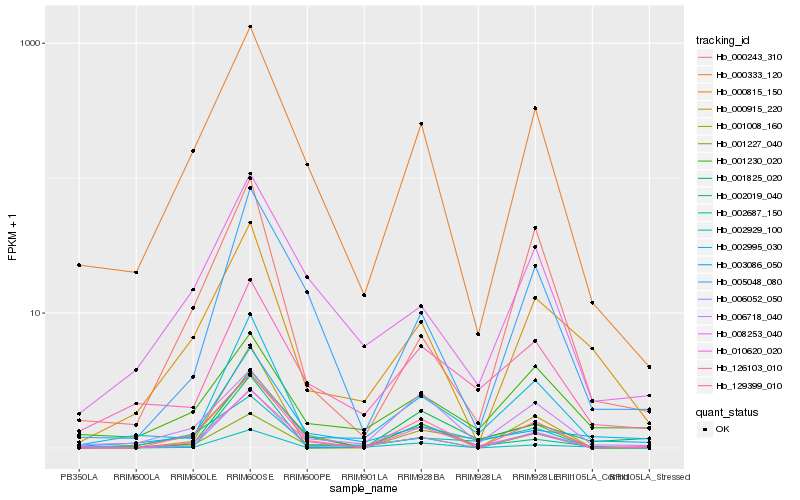
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002995_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006052_050 |
0.1584035428 |
- |
- |
hypothetical protein EUGRSUZ_A00568 [Eucalyptus grandis] |
| 3 |
Hb_126103_010 |
0.1757603105 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 4 |
Hb_001227_040 |
0.185831741 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 5 |
Hb_000333_120 |
0.1918577551 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 6 |
Hb_010620_020 |
0.1983887701 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 7 |
Hb_000815_150 |
0.2004536778 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_003086_050 |
0.2018442848 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 9 |
Hb_001008_160 |
0.2065304082 |
- |
- |
- |
| 10 |
Hb_005048_080 |
0.2104816696 |
- |
- |
PREDICTED: PAP-specific phosphatase HAL2-like [Jatropha curcas] |
| 11 |
Hb_002687_150 |
0.2162579626 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 12 |
Hb_000243_310 |
0.217492652 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 13 |
Hb_001825_020 |
0.2177748784 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_002019_040 |
0.2186581684 |
- |
- |
hypothetical protein PRUPE_ppb016160mg, partial [Prunus persica] |
| 15 |
Hb_001230_020 |
0.2200526971 |
- |
- |
hypothetical protein POPTR_0001s31610g [Populus trichocarpa] |
| 16 |
Hb_002929_100 |
0.2201954059 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 17 |
Hb_008253_040 |
0.2230998182 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 18 |
Hb_000915_220 |
0.2255648563 |
- |
- |
Lactoylglutathione lyase / glyoxalase I family protein [Theobroma cacao] |
| 19 |
Hb_129399_010 |
0.2256219632 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_006718_040 |
0.2257318486 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |