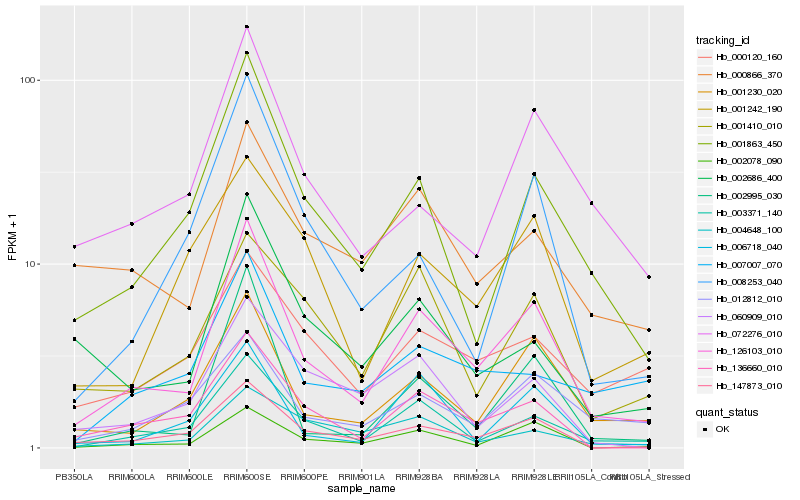
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_126103_010 |
0.0 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 2 |
Hb_003371_140 |
0.1399117949 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 3 |
Hb_147873_010 |
0.1464654775 |
- |
- |
PREDICTED: uncharacterized protein LOC102595311 [Solanum tuberosum] |
| 4 |
Hb_001230_020 |
0.1543850121 |
- |
- |
hypothetical protein POPTR_0001s31610g [Populus trichocarpa] |
| 5 |
Hb_002078_090 |
0.1553194937 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_001863_450 |
0.1585465912 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 7 |
Hb_136660_010 |
0.1641776157 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_002995_030 |
0.1757603105 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_072276_010 |
0.1853449728 |
- |
- |
PREDICTED: elongation of fatty acids protein 3-like [Jatropha curcas] |
| 10 |
Hb_012812_010 |
0.1885738814 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 11 |
Hb_006718_040 |
0.189494865 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_002686_400 |
0.1944647402 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 13 |
Hb_000866_370 |
0.1947560943 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 14 |
Hb_001410_010 |
0.1964843807 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP7-like [Jatropha curcas] |
| 15 |
Hb_008253_040 |
0.2002180656 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 16 |
Hb_060909_010 |
0.2004619954 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 17 |
Hb_001242_190 |
0.2011704201 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 18 |
Hb_004648_100 |
0.2037316613 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 19 |
Hb_007007_070 |
0.2038849618 |
transcription factor |
TF Family: TCP |
hypothetical protein MIMGU_mgv1a022855mg, partial [Erythranthe guttata] |
| 20 |
Hb_000120_160 |
0.2049957816 |
- |
- |
unnamed protein product [Vitis vinifera] |