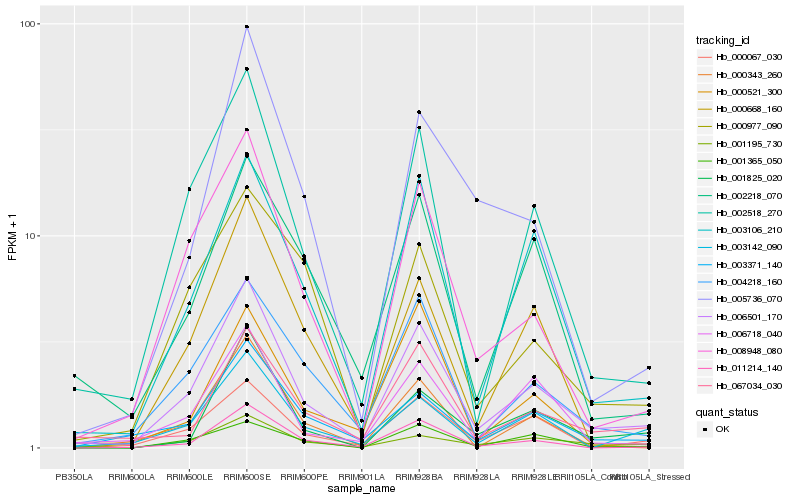
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006501_170 |
0.0 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 2 |
Hb_002518_270 |
0.1233667244 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 3 |
Hb_003106_210 |
0.1314966131 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |
| 4 |
Hb_004218_160 |
0.138564324 |
- |
- |
NBS-LRR type disease resistance protein [Medicago truncatula] |
| 5 |
Hb_008948_080 |
0.1394235842 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_003371_140 |
0.1408588448 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 7 |
Hb_011214_140 |
0.1490086262 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 8 |
Hb_000067_030 |
0.1522810856 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 9 |
Hb_003142_090 |
0.1541788712 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 10 |
Hb_067034_030 |
0.1562909752 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 11 |
Hb_006718_040 |
0.1590317217 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_001195_730 |
0.1591323044 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 13 |
Hb_001365_050 |
0.1688595731 |
- |
- |
hypothetical protein POPTR_0019s128601g, partial [Populus trichocarpa] |
| 14 |
Hb_002218_070 |
0.1713269892 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 15 |
Hb_000668_160 |
0.1778880418 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 16 |
Hb_001825_020 |
0.1789378535 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_000977_090 |
0.1797687374 |
- |
- |
hypothetical protein POPTR_0010s18550g [Populus trichocarpa] |
| 18 |
Hb_000343_260 |
0.180423377 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 19 |
Hb_005736_070 |
0.1817488733 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 20 |
Hb_000521_300 |
0.1836109214 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |