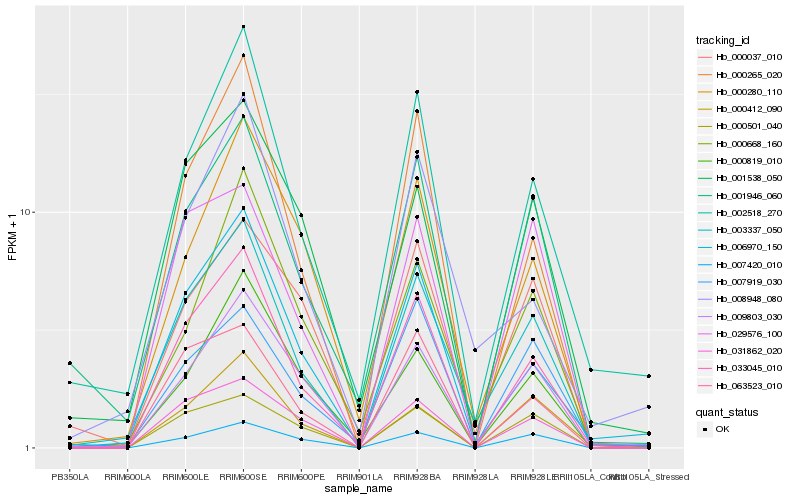
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003337_050 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 2 |
Hb_033045_010 |
0.1167831292 |
- |
- |
copine, putative [Ricinus communis] |
| 3 |
Hb_002518_270 |
0.1272142572 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 4 |
Hb_009803_030 |
0.1273207247 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 5 |
Hb_006970_150 |
0.134947184 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 6 |
Hb_001946_060 |
0.138283258 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000265_020 |
0.1477678167 |
- |
- |
hypothetical protein POPTR_0011s16980g, partial [Populus trichocarpa] |
| 8 |
Hb_008948_080 |
0.1484025384 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001538_050 |
0.1508446174 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 10 |
Hb_029576_100 |
0.1520858383 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 11 |
Hb_007420_010 |
0.1523451493 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 12 |
Hb_031862_020 |
0.1528358822 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 13 |
Hb_000412_090 |
0.1534404212 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 14 |
Hb_000280_110 |
0.1538239682 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 15 |
Hb_000668_160 |
0.1558637725 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 16 |
Hb_000501_040 |
0.158129505 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000819_010 |
0.1583404491 |
- |
- |
Receptor-like protein kinase 1, putative [Theobroma cacao] |
| 18 |
Hb_000037_010 |
0.1615293087 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |
| 19 |
Hb_063523_010 |
0.161814619 |
- |
- |
- |
| 20 |
Hb_007919_030 |
0.1622924367 |
- |
- |
- |