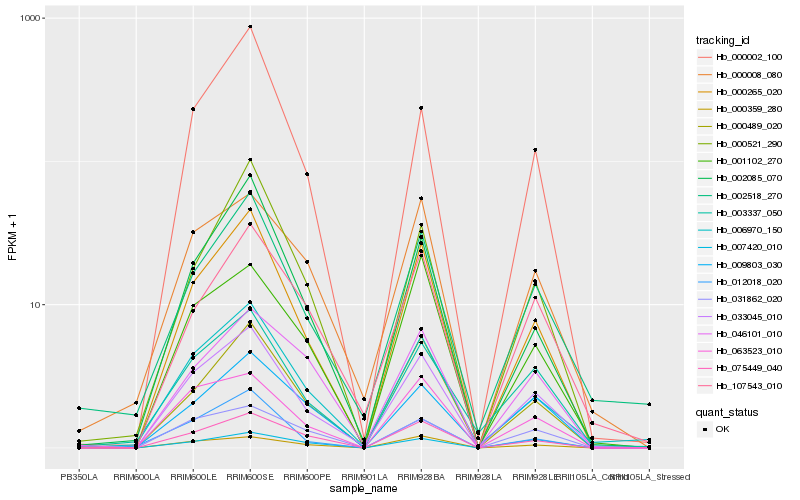
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033045_010 |
0.0 |
- |
- |
copine, putative [Ricinus communis] |
| 2 |
Hb_000265_020 |
0.078051568 |
- |
- |
hypothetical protein POPTR_0011s16980g, partial [Populus trichocarpa] |
| 3 |
Hb_006970_150 |
0.1040136004 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 4 |
Hb_063523_010 |
0.1110336508 |
- |
- |
- |
| 5 |
Hb_000489_020 |
0.1166073925 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 6 |
Hb_003337_050 |
0.1167831292 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 7 |
Hb_031862_020 |
0.1246187758 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 8 |
Hb_107543_010 |
0.1246898924 |
- |
- |
Cysteine-rich RLK 10, putative [Theobroma cacao] |
| 9 |
Hb_002518_270 |
0.128727666 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 10 |
Hb_000359_280 |
0.130674772 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |
| 11 |
Hb_075449_040 |
0.1307139007 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-1 [Jatropha curcas] |
| 12 |
Hb_000008_080 |
0.1313455398 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000002_100 |
0.1348126212 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 14 |
Hb_009803_030 |
0.1359857464 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 15 |
Hb_001102_270 |
0.1361434325 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 16 |
Hb_012018_020 |
0.1383583064 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Eucalyptus grandis] |
| 17 |
Hb_002085_070 |
0.1413555311 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_046101_010 |
0.1452396422 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 19 |
Hb_000521_290 |
0.1457610604 |
- |
- |
hypothetical protein POPTR_0001s23530g [Populus trichocarpa] |
| 20 |
Hb_007420_010 |
0.1458694288 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |