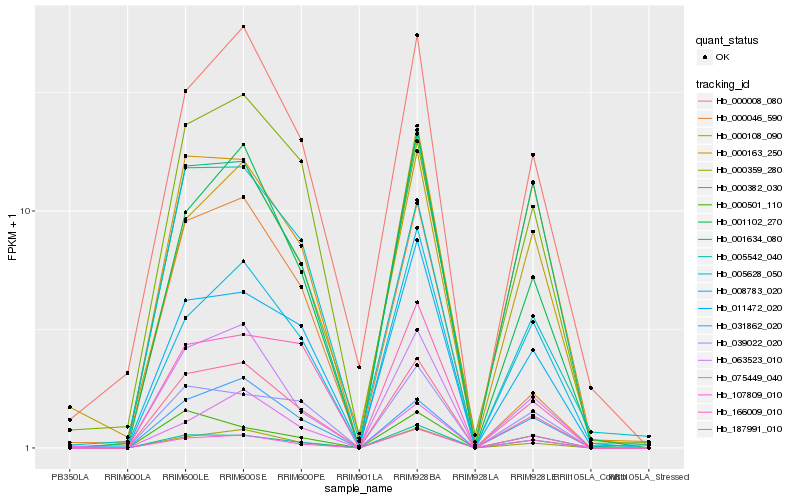
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_187991_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 2 |
Hb_000359_280 |
0.0618559628 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |
| 3 |
Hb_063523_010 |
0.0697762596 |
- |
- |
- |
| 4 |
Hb_001102_270 |
0.0995981342 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 5 |
Hb_031862_020 |
0.107347143 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 6 |
Hb_005542_040 |
0.1127410636 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 7 |
Hb_000008_080 |
0.1170193972 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_107809_010 |
0.1195196481 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 9 |
Hb_011472_020 |
0.1196587949 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |
| 10 |
Hb_039022_020 |
0.122020289 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 11 |
Hb_000046_590 |
0.1285689894 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 12 |
Hb_000108_090 |
0.1295973273 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 13 |
Hb_166009_010 |
0.1310097759 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 14 |
Hb_005628_050 |
0.1337699842 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_008783_020 |
0.1356263246 |
- |
- |
hypothetical protein JCGZ_21284 [Jatropha curcas] |
| 16 |
Hb_075449_040 |
0.1365246559 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-1 [Jatropha curcas] |
| 17 |
Hb_000382_030 |
0.1366597999 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 18 |
Hb_000163_250 |
0.1368325542 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_001634_080 |
0.1373332079 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_000501_110 |
0.1383181687 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 [Populus euphratica] |