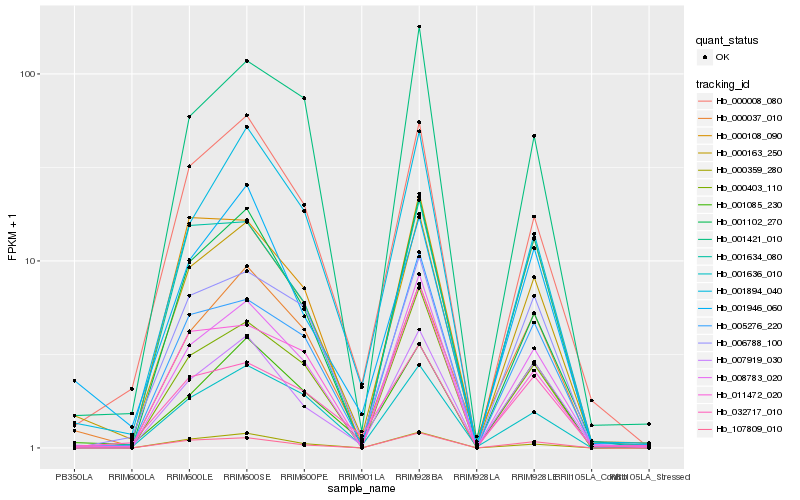
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000163_250 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_007919_030 |
0.0862271405 |
- |
- |
- |
| 3 |
Hb_000008_080 |
0.0956300045 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_008783_020 |
0.1057217881 |
- |
- |
hypothetical protein JCGZ_21284 [Jatropha curcas] |
| 5 |
Hb_000037_010 |
0.1061848052 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |
| 6 |
Hb_005276_220 |
0.1088272974 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 7 |
Hb_006788_100 |
0.1113850629 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000403_110 |
0.1119238011 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 9 |
Hb_107809_010 |
0.1149323631 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 10 |
Hb_001946_060 |
0.1163951437 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001894_040 |
0.1177019655 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001085_230 |
0.1179898322 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 13 |
Hb_032717_010 |
0.119011176 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_001636_010 |
0.1236286638 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 15 |
Hb_001421_010 |
0.1252741281 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000359_280 |
0.1259818314 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |
| 17 |
Hb_000108_090 |
0.1265942784 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 18 |
Hb_001102_270 |
0.1275533968 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 19 |
Hb_001634_080 |
0.128137942 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_011472_020 |
0.1290139464 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |