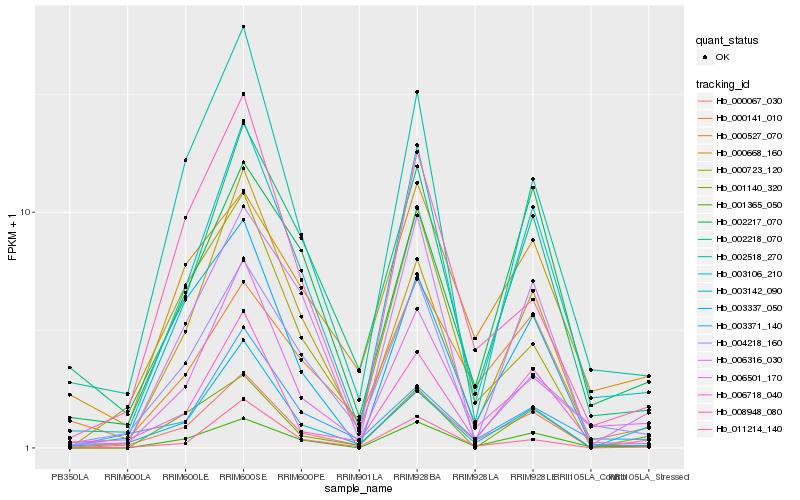
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000067_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 2 |
Hb_003106_210 |
0.1323303886 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |
| 3 |
Hb_003142_090 |
0.1326288822 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 4 |
Hb_001365_050 |
0.1432244622 |
- |
- |
hypothetical protein POPTR_0019s128601g, partial [Populus trichocarpa] |
| 5 |
Hb_006501_170 |
0.1522810856 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 6 |
Hb_006718_040 |
0.153302813 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_001140_320 |
0.1596513643 |
- |
- |
PREDICTED: oxalate--CoA ligase [Jatropha curcas] |
| 8 |
Hb_006316_030 |
0.1659426618 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 9 |
Hb_011214_140 |
0.1695319135 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 10 |
Hb_002218_070 |
0.1747753727 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 11 |
Hb_002518_270 |
0.177049287 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 12 |
Hb_003337_050 |
0.1775692548 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 13 |
Hb_000141_010 |
0.1780851559 |
- |
- |
PREDICTED: uncharacterized protein LOC105632253 [Jatropha curcas] |
| 14 |
Hb_003371_140 |
0.185109901 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 15 |
Hb_004218_160 |
0.1851581605 |
- |
- |
NBS-LRR type disease resistance protein [Medicago truncatula] |
| 16 |
Hb_008948_080 |
0.1855535581 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000668_160 |
0.1857984213 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 18 |
Hb_002217_070 |
0.1859949131 |
- |
- |
PREDICTED: uncharacterized protein LOC105643596 [Jatropha curcas] |
| 19 |
Hb_000527_070 |
0.1860139825 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 20 |
Hb_000723_120 |
0.1863685632 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A1 [Jatropha curcas] |