| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
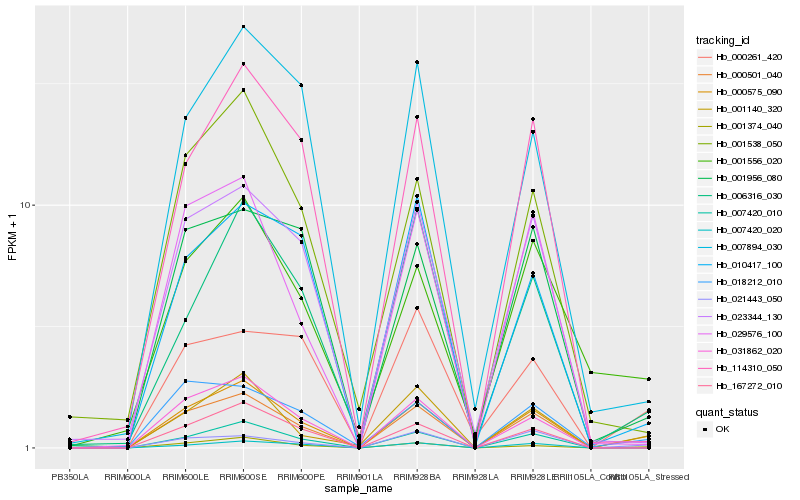
Hb_000575_090 |
0.0 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 24 [Populus euphratica] |
| 2 |
Hb_001374_040 |
0.1070563161 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Gossypium raimondii] |
| 3 |
Hb_001140_320 |
0.1339945654 |
- |
- |
PREDICTED: oxalate--CoA ligase [Jatropha curcas] |
| 4 |
Hb_018212_010 |
0.1491769148 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 5 |
Hb_000501_040 |
0.1520137657 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_007420_010 |
0.1562495869 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 7 |
Hb_031862_020 |
0.1601998142 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 8 |
Hb_167272_010 |
0.1608124746 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 9 |
Hb_006316_030 |
0.1708993444 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 10 |
Hb_001556_020 |
0.1716045308 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 11 |
Hb_114310_050 |
0.1720649092 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 12 |
Hb_021443_050 |
0.1735866321 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_010417_100 |
0.1741062272 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 14 |
Hb_023344_130 |
0.1741811431 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_029576_100 |
0.1746718187 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 16 |
Hb_001956_080 |
0.1751131291 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 17 |
Hb_001538_050 |
0.1759319699 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 18 |
Hb_000261_420 |
0.1766924564 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 19 |
Hb_007420_020 |
0.1789697605 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 20 |
Hb_007894_030 |
0.1790898887 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |