| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
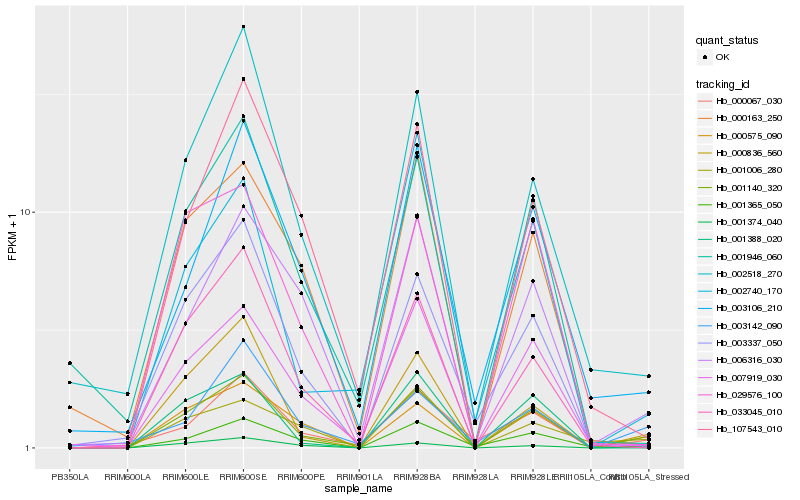
Hb_001140_320 |
0.0 |
- |
- |
PREDICTED: oxalate--CoA ligase [Jatropha curcas] |
| 2 |
Hb_000575_090 |
0.1339945654 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 24 [Populus euphratica] |
| 3 |
Hb_000836_560 |
0.144160211 |
- |
- |
- |
| 4 |
Hb_000067_030 |
0.1596513643 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 5 |
Hb_001374_040 |
0.1675169081 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Gossypium raimondii] |
| 6 |
Hb_007919_030 |
0.16910028 |
- |
- |
- |
| 7 |
Hb_001365_050 |
0.1705057461 |
- |
- |
hypothetical protein POPTR_0019s128601g, partial [Populus trichocarpa] |
| 8 |
Hb_006316_030 |
0.1709295847 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 9 |
Hb_033045_010 |
0.1788548114 |
- |
- |
copine, putative [Ricinus communis] |
| 10 |
Hb_002518_270 |
0.1797161214 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 11 |
Hb_001946_060 |
0.1813470683 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_003106_210 |
0.1843815545 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |
| 13 |
Hb_001388_020 |
0.188109847 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP20 [Jatropha curcas] |
| 14 |
Hb_107543_010 |
0.1888425044 |
- |
- |
Cysteine-rich RLK 10, putative [Theobroma cacao] |
| 15 |
Hb_001006_280 |
0.1888604266 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 16 |
Hb_003337_050 |
0.1931462499 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 17 |
Hb_002740_170 |
0.1936189852 |
- |
- |
PREDICTED: uncharacterized protein LOC105647357 [Jatropha curcas] |
| 18 |
Hb_003142_090 |
0.193807399 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 19 |
Hb_029576_100 |
0.1942684584 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 20 |
Hb_000163_250 |
0.1950114544 |
- |
- |
unnamed protein product [Vitis vinifera] |