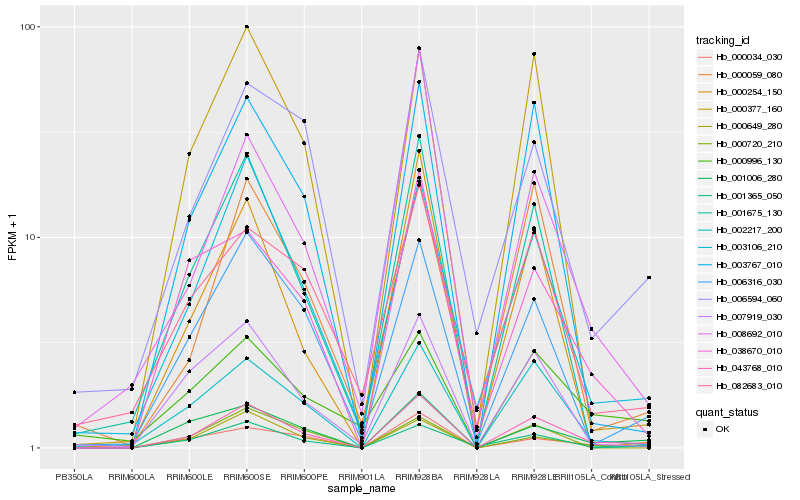
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_043768_010 |
0.0 |
- |
- |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 2 |
Hb_002217_200 |
0.1320051658 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 3 |
Hb_001365_050 |
0.1376401551 |
- |
- |
hypothetical protein POPTR_0019s128601g, partial [Populus trichocarpa] |
| 4 |
Hb_001006_280 |
0.1446198163 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 5 |
Hb_000034_030 |
0.1453319206 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 6 |
Hb_003106_210 |
0.1475003833 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |
| 7 |
Hb_003767_010 |
0.1568754099 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 8 |
Hb_006316_030 |
0.1618342194 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 9 |
Hb_038670_010 |
0.1730862554 |
- |
- |
Major allergen Pru ar, putative [Ricinus communis] |
| 10 |
Hb_006594_060 |
0.1733087709 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 11 |
Hb_007919_030 |
0.1765525892 |
- |
- |
- |
| 12 |
Hb_001675_130 |
0.1779600708 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 13 |
Hb_000720_210 |
0.1805139672 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 14 |
Hb_000059_080 |
0.1814173945 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 15 |
Hb_000377_160 |
0.1832024215 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 16 |
Hb_000254_150 |
0.1849268478 |
- |
- |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 17 |
Hb_000649_280 |
0.1880536087 |
- |
- |
PREDICTED: F-box protein PP2-B10-like [Pyrus x bretschneideri] |
| 18 |
Hb_008692_010 |
0.1890077373 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 19 |
Hb_082683_010 |
0.1892800534 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_000996_130 |
0.1892906766 |
- |
- |
PREDICTED: remorin-like [Populus euphratica] |