| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
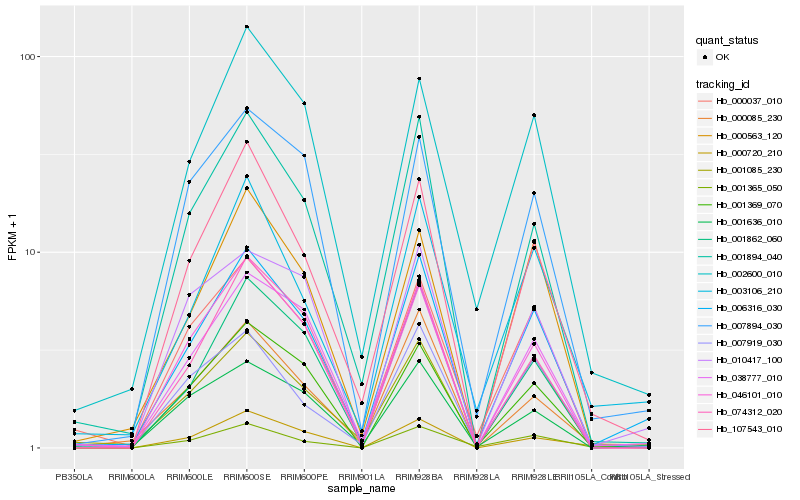
Hb_006316_030 |
0.0 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 2 |
Hb_038777_010 |
0.1030450053 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |
| 3 |
Hb_003106_210 |
0.1101219667 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |
| 4 |
Hb_001862_060 |
0.1164712855 |
- |
- |
PREDICTED: uncharacterized protein LOC105639802 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001894_040 |
0.1167935561 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001636_010 |
0.117525775 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 7 |
Hb_001085_230 |
0.1206117572 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 8 |
Hb_001365_050 |
0.1222291709 |
- |
- |
hypothetical protein POPTR_0019s128601g, partial [Populus trichocarpa] |
| 9 |
Hb_107543_010 |
0.1229583443 |
- |
- |
Cysteine-rich RLK 10, putative [Theobroma cacao] |
| 10 |
Hb_046101_010 |
0.1270698782 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 11 |
Hb_000037_010 |
0.1276815017 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |
| 12 |
Hb_002600_010 |
0.128799284 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_007894_030 |
0.1294647273 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 14 |
Hb_000720_210 |
0.1294926194 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 15 |
Hb_000563_120 |
0.1310825072 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 16 |
Hb_001369_070 |
0.1316682833 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000085_230 |
0.1327428784 |
- |
- |
PREDICTED: probable long-chain-alcohol O-fatty-acyltransferase 5 [Jatropha curcas] |
| 18 |
Hb_010417_100 |
0.1328097601 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 19 |
Hb_007919_030 |
0.1360930164 |
- |
- |
- |
| 20 |
Hb_074312_020 |
0.1387597843 |
- |
- |
cytochrome P450, putative [Ricinus communis] |