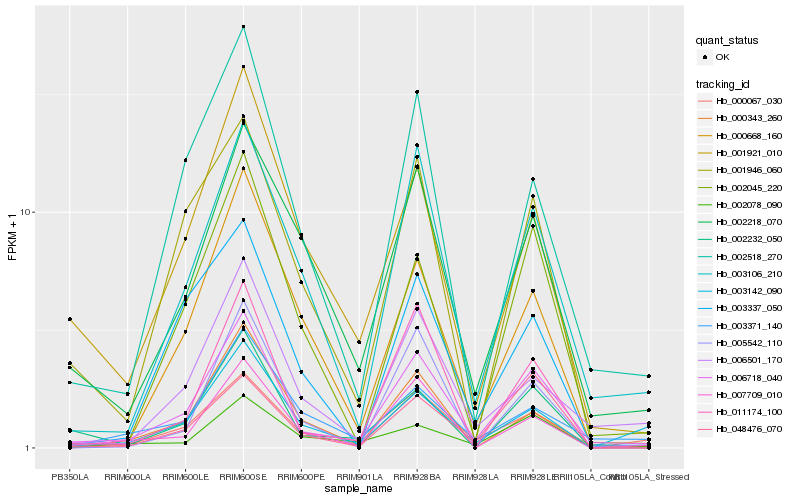
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006718_040 |
0.0 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_003106_210 |
0.1369083522 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |
| 3 |
Hb_000067_030 |
0.153302813 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 4 |
Hb_002518_270 |
0.1544239331 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 5 |
Hb_011174_100 |
0.1547983262 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_006501_170 |
0.1590317217 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 7 |
Hb_002218_070 |
0.1634368175 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 8 |
Hb_002045_220 |
0.1705197519 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002232_050 |
0.171244015 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 10 |
Hb_007709_010 |
0.1717148128 |
- |
- |
- |
| 11 |
Hb_001946_060 |
0.1722031633 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001921_010 |
0.1783512827 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 13 |
Hb_003371_140 |
0.1809832801 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 14 |
Hb_002078_090 |
0.1844585921 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000668_160 |
0.1846409741 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 16 |
Hb_005542_110 |
0.1855960868 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 17 |
Hb_003142_090 |
0.1865637574 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 18 |
Hb_000343_260 |
0.1874979941 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 19 |
Hb_003337_050 |
0.1879744043 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 20 |
Hb_048476_070 |
0.1880316246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |