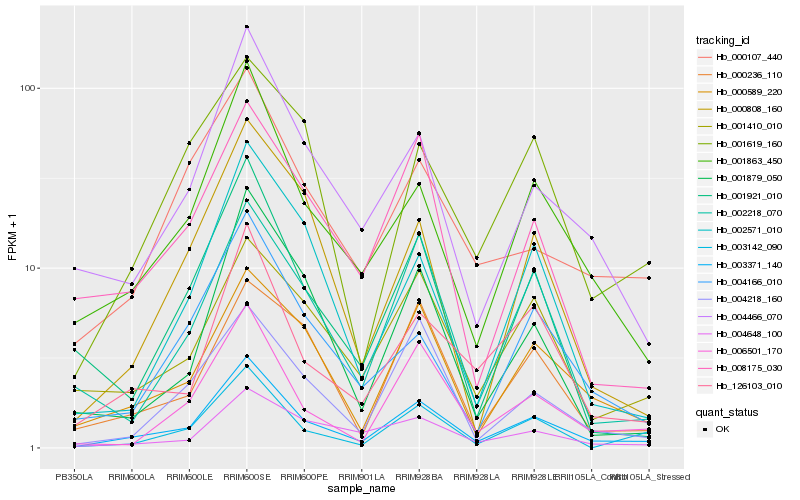
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003371_140 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 2 |
Hb_001863_450 |
0.1225703635 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 3 |
Hb_126103_010 |
0.1399117949 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 4 |
Hb_006501_170 |
0.1408588448 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 5 |
Hb_000589_220 |
0.1417453513 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 6 |
Hb_004466_070 |
0.1444752198 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 7 |
Hb_003142_090 |
0.1540182335 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 8 |
Hb_004166_010 |
0.1609872646 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 9 |
Hb_001410_010 |
0.1627744506 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP7-like [Jatropha curcas] |
| 10 |
Hb_000107_440 |
0.1642338587 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 11 |
Hb_004218_160 |
0.1645928063 |
- |
- |
NBS-LRR type disease resistance protein [Medicago truncatula] |
| 12 |
Hb_002218_070 |
0.1646372257 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 13 |
Hb_001921_010 |
0.1649694432 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 14 |
Hb_004648_100 |
0.1652081147 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 15 |
Hb_001879_050 |
0.1696097717 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 16 |
Hb_001619_160 |
0.1702012743 |
transcription factor |
TF Family: ERF |
DREB1p [Hevea brasiliensis] |
| 17 |
Hb_008175_030 |
0.1704308622 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |
| 18 |
Hb_002571_010 |
0.1706301702 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_000808_160 |
0.1726556852 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 20 |
Hb_000236_110 |
0.1745524129 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic [Jatropha curcas] |