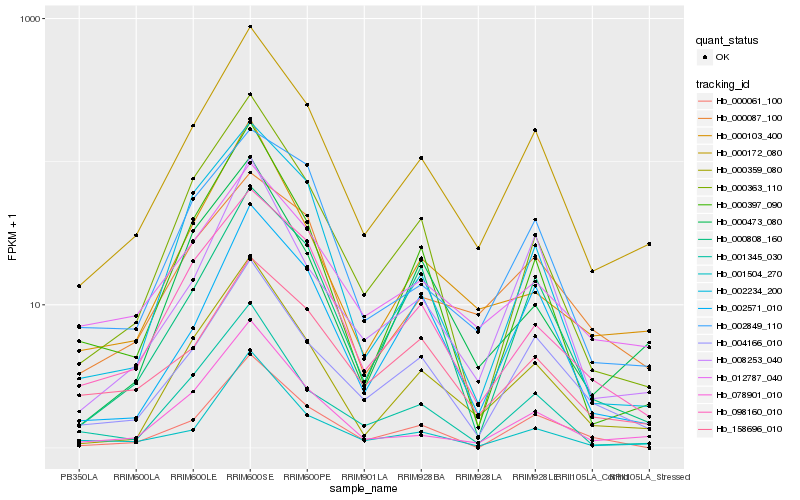
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_080 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 2 |
Hb_000359_080 |
0.1016928606 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 3 |
Hb_004166_010 |
0.1046915161 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 4 |
Hb_078901_010 |
0.1177727918 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 5 |
Hb_008253_040 |
0.1218217007 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 6 |
Hb_001345_030 |
0.1246679387 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |
| 7 |
Hb_098160_010 |
0.1264102177 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000363_110 |
0.1283675831 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 21 [Jatropha curcas] |
| 9 |
Hb_002571_010 |
0.12917642 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_012787_040 |
0.1297809789 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000087_100 |
0.1326655432 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 12 |
Hb_002849_110 |
0.1346909931 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 13 |
Hb_158696_010 |
0.1369879207 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 14 |
Hb_000808_160 |
0.1371363594 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 15 |
Hb_000061_100 |
0.1385016553 |
- |
- |
hypothetical protein CISIN_1g0053742mg, partial [Citrus sinensis] |
| 16 |
Hb_001504_270 |
0.1385179132 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 17 |
Hb_000103_400 |
0.139815375 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000473_080 |
0.1418744084 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 19 |
Hb_000397_090 |
0.143115709 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 20 |
Hb_002234_200 |
0.1433097349 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |