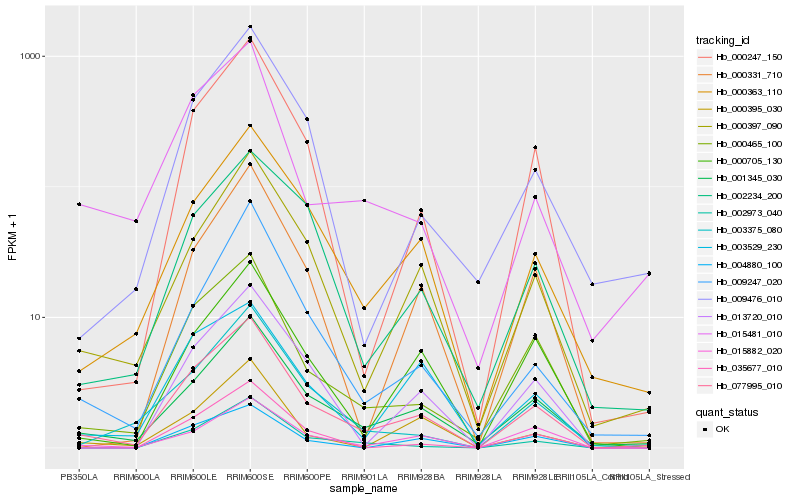
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_077995_010 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 2 |
Hb_001345_030 |
0.1009021917 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |
| 3 |
Hb_000465_100 |
0.1159751849 |
- |
- |
catalytic, putative [Ricinus communis] |
| 4 |
Hb_000397_090 |
0.1331993696 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 5 |
Hb_000363_110 |
0.1344866235 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 21 [Jatropha curcas] |
| 6 |
Hb_000247_150 |
0.1435576275 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 7 |
Hb_013720_010 |
0.1555994726 |
- |
- |
Putative DNA repair RAD23-3 -like protein [Gossypium arboreum] |
| 8 |
Hb_000705_130 |
0.1579193359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002973_040 |
0.1604613112 |
- |
- |
hypothetical protein CICLE_v10020565mg [Citrus clementina] |
| 10 |
Hb_015882_020 |
0.1607534639 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 11 |
Hb_000331_710 |
0.1617226027 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_009247_020 |
0.1628026474 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 13 |
Hb_000395_030 |
0.1634764638 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 14 |
Hb_003529_230 |
0.1648480215 |
transcription factor |
TF Family: ERF |
- |
| 15 |
Hb_015481_010 |
0.165622531 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |
| 16 |
Hb_003375_080 |
0.1665020908 |
- |
- |
PREDICTED: uncharacterized protein LOC105643382 isoform X1 [Jatropha curcas] |
| 17 |
Hb_009476_010 |
0.1701705285 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 18 |
Hb_035677_010 |
0.170614394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002234_200 |
0.1707142375 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 20 |
Hb_004880_100 |
0.1707313961 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300-like [Glycine max] |