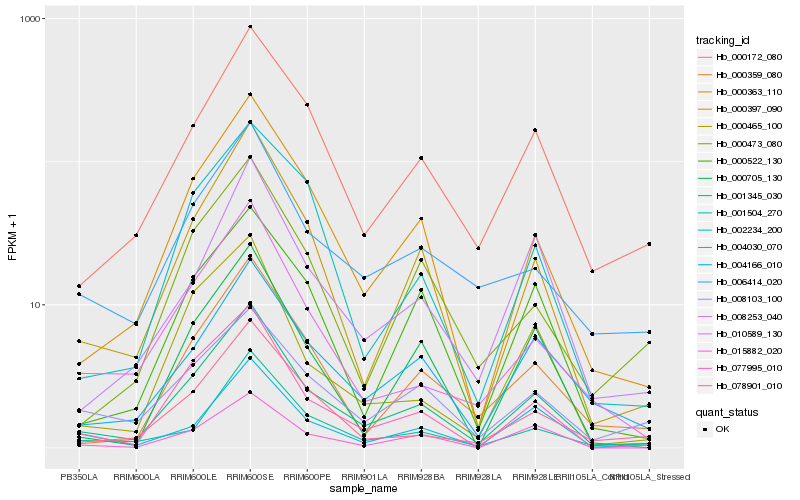
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001345_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |
| 2 |
Hb_000397_090 |
0.0970723259 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 3 |
Hb_000363_110 |
0.098281347 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 21 [Jatropha curcas] |
| 4 |
Hb_077995_010 |
0.1009021917 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 5 |
Hb_000172_080 |
0.1246679387 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 6 |
Hb_004166_010 |
0.1339360587 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 7 |
Hb_000359_080 |
0.1350907093 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 8 |
Hb_015882_020 |
0.1371384612 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 9 |
Hb_001504_270 |
0.1393697689 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 10 |
Hb_008253_040 |
0.1429937292 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 11 |
Hb_004030_070 |
0.1434081772 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 12 |
Hb_078901_010 |
0.1437646979 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 13 |
Hb_010589_130 |
0.1447205249 |
- |
- |
PREDICTED: uncharacterized protein LOC104884346 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_000465_100 |
0.1471638783 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_000705_130 |
0.1486742628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006414_020 |
0.1497577921 |
transcription factor |
TF Family: MYB |
- |
| 17 |
Hb_002234_200 |
0.1508592868 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 18 |
Hb_000522_130 |
0.1515006772 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 19 |
Hb_008103_100 |
0.1520427986 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 20 |
Hb_000473_080 |
0.1564831744 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |